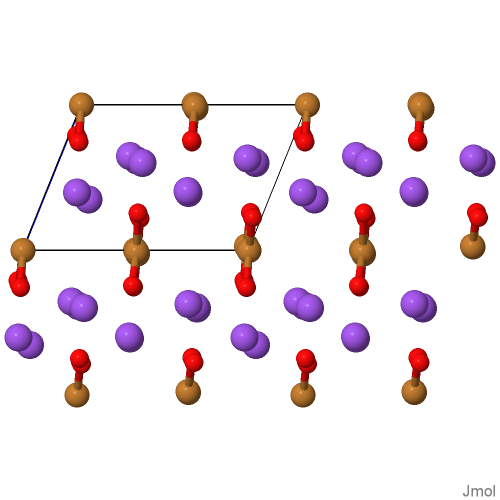
Structures of incommensurate and commensurate composite crystals NaxCuO2 (x = 1.58, 1.6, 1.62)
Authors:van Smaalen, Sander; Dinnebier, Robert ; Sofin, Mikhail; Jansen,Martin
Journal:Acta Crystallographica, Section B 63 17-25 (2007)
DOI:https://doi.org/10.1107/S0108768106039462
B-IncStrDB ID: N9lhzei9O8t Entry date: 2022-01-28 Last revision: 2022-01-28
I
Chemical data
Full Name: sodium copper oxide [ Help ]
Structural Formula Sum: Cu1 Na1.57664 O2 [ Help ]
Formula weight: 661.6 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Superspace group name: C2/m(0β0)s0 [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,x2,-x3,1/2+x4 |
| 3 | -x1,-x2,-x3,-x4 |
| 4 | x1,-x2,x3,1/2-x4 |
| 5 | 1/2+x1,1/2+x2,x3,x4 |
| 6 | 1/2-x1,1/2+x2,-x3,1/2+x4 |
| 7 | 1/2-x1,1/2-x2,-x3,-x4 |
| 8 | 1/2+x1,1/2-x2,x3,1/2-x4 |
a: 8.24153(4) Å [ Help ]
b: 2.78589(2) Å [ Help ]
c: 5.71687(3) Å [ Help ]
α: 90 ° [ Help ]
β: 111.9698(5) ° [ Help ]
γ: 90 ° [ Help ]
Volume: 121.7270(10) Å3 [ Help ]
Z: 2 [ Help ]
Cell measurement temperature: 293 K [ Help ]
Modulation dimension: 1 [ Help ]
Number of subsystems: 2 [ Help ]
Subsystem code: 1 [ Help ]
Subsystem description: 1-st subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
Subsystem code: 2 [ Help ]
Subsystem description: 2-nd subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.78832(2) | 0.00000 |
μ: 17.561 mm-1 [ Help ]
Absorption correction type: none [ Help ]
Absorption correction remarks: (Jana2000; Petricek, Dusek & Palatinus, 2000) [ Help ]
Refinement details
R(all): 0.0482 [ Help ]
R(obs): 0.0468 [ Help ]
wR(obs): 0.0454 [ Help ]
wR(all): 0.0455 [ Help ]
Nb. of parameters: 36 [ Help ]
Number of restraints: 0 [ Help ]
Weighting scheme: sigma [ Help ]
Δ/σ(max): 0.2946 [ Help ]
Δ/σ(mean): 0.0323 [ Help ]
Extinction method: none [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom symbol | Atom site label | Subsystem | x | y | z | Uiso/equiv | ADP type | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Occupancy | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu | Cu1 | 1 | 0 | 0.5 | 0 | 0.0038(2) | Uiso | d | . | 1 | . | . |
| O | O1 | 1 | 0.0493(3) | 0 | 0.2416(3) | 0.0091(7) | Uiso | d | . | 1 | . | . |
| Na | Na1 | 2 | -0.1512(3) | 0.25 | 0.3832(4) | 0.0141(6) | Uiso | d | . | 1 | . | . |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.78832 | 0.00000 |
| 2 | 0.00000 | 1.57664 | 0.00000 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Cu1x1 | Cu1 | x | 1 |
| Cu1y1 | Cu1 | y | 1 |
| Cu1z1 | Cu1 | z | 1 |
| Cu1x2 | Cu1 | x | 2 |
| Cu1y2 | Cu1 | y | 2 |
| Cu1z2 | Cu1 | z | 2 |
| O1x1 | O1 | x | 1 |
| O1y1 | O1 | y | 1 |
| O1z1 | O1 | z | 1 |
| O1x2 | O1 | x | 2 |
| O1y2 | O1 | y | 2 |
| O1z2 | O1 | z | 2 |
| Na1x1 | Na1 | x | 1 |
| Na1y1 | Na1 | y | 1 |
| Na1z1 | Na1 | z | 1 |
| Na1x2 | Na1 | x | 2 |
| Na1y2 | Na1 | y | 2 |
| Na1z2 | Na1 | z | 2 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cu1x1 | 0 | -0.01696(19) |
| Cu1y1 | 0 | 0 |
| Cu1z1 | 0 | -0.0291(3) |
| Cu1x2 | 0 | 0 |
| Cu1y2 | 0 | -0.0041(10) |
| Cu1z2 | 0 | 0 |
| O1x1 | 0 | 0.0117(7) |
| O1y1 | 0.0323(19) | 0 |
| O1z1 | 0 | 0.0232(8) |
| O1x2 | 0.0037(12) | 0 |
| O1y2 | 0 | 0.021(3) |
| O1z2 | 0.0074(18) | 0 |
| Na1x1 | -0.0358(5) | 0 |
| Na1y1 | 0 | 0.1111(9) |
| Na1z1 | -0.0233(7) | 0 |
| Na1x2 | -0.0201(9) | 0 |
| Na1y2 | 0 | 0.0160(15) |
| Na1z2 | -0.0177(12) | 0 |
II
Chemical data
Structural Formula Sum: Cu5 Na8 O10 [ Help ]
Formula weight: 661.6 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Superspace group name: C2/m(0β0)s0 [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,x2,-x3,1/2+x4 |
| 3 | -x1,-x2,-x3,-x4 |
| 4 | x1,-x2,x3,1/2-x4 |
| 5 | 1/2+x1,1/2+x2,x3,x4 |
| 6 | 1/2-x1,1/2+x2,-x3,1/2+x4 |
| 7 | 1/2-x1,1/2-x2,-x3,-x4 |
| 8 | 1/2+x1,1/2-x2,x3,1/2-x4 |
a: 8.228(1) Å [ Help ]
b: 2.7858(4) Å [ Help ]
c: 5.707(1) Å [ Help ]
α: 90 ° [ Help ]
β: 111.718(2) ° [ Help ]
γ: 90 ° [ Help ]
Volume: 121.528 Å3 [ Help ]
Z: 2 [ Help ]
Modulation dimension: 1 [ Help ]
Number of subsystems: 2 [ Help ]
Subsystem code: 1 [ Help ]
Subsystem description: 1-st subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
Subsystem code: 2 [ Help ]
Subsystem description: 2-nd subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.80000 | 0.00000 |
μ: 22.317 mm-1 [ Help ]
Absorption correction type: none [ Help ]
Minimum transmission factor: 1.000 [ Help ]
Maximum transmission factor: 1.000 [ Help ]
Refinement details
Total nb. of reflections: 5656 [ Help ]
Nb. of observed reflections: 4523 [ Help ]
Intense reflections threshold: I>3σ(I) [ Help ]
Refinement based on: F [ Help ]
R(all): 0.0418 [ Help ]
R(obs): 0.0347 [ Help ]
wR(obs): 0.0332 [ Help ]
wR(all): 0.0334 [ Help ]
S(all): 2.95 [ Help ]
Nb. of reflections: 5656 [ Help ]
Nb. of parameters: 107 [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/σ2(F) [ Help ]
Δ/σ(max): 0.0018 [ Help ]
Δ/σ(mean): 0.0002 [ Help ]
Refinement remarks: Dubious amplitudes of some ADP modulations. [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom symbol | Atom site label | Subsystem | x | y | z | Uiso/equiv | ADP type | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Occupancy | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu | Cu1 | 1 | 0 | 0.5 | 0 | 0.0107 | Uani | d | . | 1 | . | . |
| O | O1 | 1 | 0.05204 | 0 | 0.242277 | 0.0161 | Uani | d | . | 1 | . | . |
| Na | Na1 | 2 | -0.154182 | 0.25 | 0.377855 | 0.0268 | Uani | d | . | 1 | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U12 | U13 | U23 | Atom site symbol |
|---|---|---|---|---|---|---|---|
| Cu1 | 0.012659 | 0.010929 | 0.008165 | 0 | 0.003339 | 0 | Cu |
| O1 | 0.023003 | 0.014862 | 0.009429 | 0 | 0.004993 | 0 | O |
| Na1 | 0.021659 | 0.042864 | 0.016004 | 0 | 0.007236 | 0 | Na |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.80000 | 0.00000 |
| 2 | 0.00000 | 1.60000 | 0.00000 |
| 3 | 0.00000 | 2.40000 | 0.00000 |
| 4 | 0.00000 | 3.20000 | 0.00000 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Cu1x1 | Cu1 | x | 1 |
| Cu1y1 | Cu1 | y | 1 |
| Cu1z1 | Cu1 | z | 1 |
| Cu1x2 | Cu1 | x | 2 |
| Cu1y2 | Cu1 | y | 2 |
| Cu1z2 | Cu1 | z | 2 |
| Cu1x3 | Cu1 | x | 3 |
| Cu1y3 | Cu1 | y | 3 |
| Cu1z3 | Cu1 | z | 3 |
| Cu1x4 | Cu1 | x | 4 |
| Cu1y4 | Cu1 | y | 4 |
| Cu1z4 | Cu1 | z | 4 |
| O1x1 | O1 | x | 1 |
| O1y1 | O1 | y | 1 |
| O1z1 | O1 | z | 1 |
| O1x2 | O1 | x | 2 |
| O1y2 | O1 | y | 2 |
| O1z2 | O1 | z | 2 |
| O1x3 | O1 | x | 3 |
| O1y3 | O1 | y | 3 |
| O1z3 | O1 | z | 3 |
| O1x4 | O1 | x | 4 |
| O1y4 | O1 | y | 4 |
| O1z4 | O1 | z | 4 |
| Na1x1 | Na1 | x | 1 |
| Na1y1 | Na1 | y | 1 |
| Na1z1 | Na1 | z | 1 |
| Na1x2 | Na1 | x | 2 |
| Na1y2 | Na1 | y | 2 |
| Na1z2 | Na1 | z | 2 |
| Na1x3 | Na1 | x | 3 |
| Na1y3 | Na1 | y | 3 |
| Na1z3 | Na1 | z | 3 |
| Na1x4 | Na1 | x | 4 |
| Na1y4 | Na1 | y | 4 |
| Na1z4 | Na1 | z | 4 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cu1x1 | 0 | -0.0143 |
| Cu1y1 | 0 | 0 |
| Cu1z1 | 0 | -0.0286 |
| Cu1x2 | 0 | 0 |
| Cu1y2 | 0 | -0.0062 |
| Cu1z2 | 0 | 0 |
| Cu1x3 | 0 | 0.0042 |
| Cu1y3 | 0 | 0 |
| Cu1z3 | 0 | 0.0027 |
| Cu1x4 | 0 | 0 |
| Cu1y4 | 0 | -0.0014 |
| Cu1z4 | 0 | 0 |
| O1x1 | 0 | 0.0119 |
| O1y1 | 0.0239 | 0 |
| O1z1 | 0 | 0.0243 |
| O1x2 | -0.0061 | 0 |
| O1y2 | 0 | 0.0204 |
| O1z2 | -0.0075 | 0 |
| O1x3 | 0 | 0.0076 |
| O1y3 | -0.0082 | 0 |
| O1z3 | 0 | 0.0043 |
| O1x4 | 0.0013 | 0 |
| O1y4 | 0 | 0.0048 |
| O1z4 | 0.0003 | 0 |
| Na1x1 | -0.0364 | 0 |
| Na1y1 | 0 | 0.0985 |
| Na1z1 | -0.0256 | 0 |
| Na1x2 | -0.0082 | 0 |
| Na1y2 | 0 | -0.0018 |
| Na1z2 | -0.0097 | 0 |
| Na1x3 | 0.0042 | 0 |
| Na1y3 | 0 | -0.0179 |
| Na1z3 | -0.0007 | 0 |
| Na1x4 | 0.0024 | 0 |
| Na1y4 | 0 | 0 |
| Na1z4 | 0.0004 | 0 |
Definition of the ADP Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Tensor element | Wave vector code |
|---|---|---|---|
| Cu1U111 | Cu1 | U11 | 1 |
| Cu1U221 | Cu1 | U22 | 1 |
| Cu1U331 | Cu1 | U33 | 1 |
| Cu1U121 | Cu1 | U12 | 1 |
| Cu1U131 | Cu1 | U13 | 1 |
| Cu1U231 | Cu1 | U23 | 1 |
| Cu1U112 | Cu1 | U11 | 2 |
| Cu1U222 | Cu1 | U22 | 2 |
| Cu1U332 | Cu1 | U33 | 2 |
| Cu1U122 | Cu1 | U12 | 2 |
| Cu1U132 | Cu1 | U13 | 2 |
| Cu1U232 | Cu1 | U23 | 2 |
| Cu1U113 | Cu1 | U11 | 3 |
| Cu1U223 | Cu1 | U22 | 3 |
| Cu1U333 | Cu1 | U33 | 3 |
| Cu1U123 | Cu1 | U12 | 3 |
| Cu1U133 | Cu1 | U13 | 3 |
| Cu1U233 | Cu1 | U23 | 3 |
| Cu1U114 | Cu1 | U11 | 4 |
| Cu1U224 | Cu1 | U22 | 4 |
| Cu1U334 | Cu1 | U33 | 4 |
| Cu1U124 | Cu1 | U12 | 4 |
| Cu1U134 | Cu1 | U13 | 4 |
| Cu1U234 | Cu1 | U23 | 4 |
| O1U111 | O1 | U11 | 1 |
| O1U221 | O1 | U22 | 1 |
| O1U331 | O1 | U33 | 1 |
| O1U121 | O1 | U12 | 1 |
| O1U131 | O1 | U13 | 1 |
| O1U231 | O1 | U23 | 1 |
| O1U112 | O1 | U11 | 2 |
| O1U222 | O1 | U22 | 2 |
| O1U332 | O1 | U33 | 2 |
| O1U122 | O1 | U12 | 2 |
| O1U132 | O1 | U13 | 2 |
| O1U232 | O1 | U23 | 2 |
| O1U113 | O1 | U11 | 3 |
| O1U223 | O1 | U22 | 3 |
| O1U333 | O1 | U33 | 3 |
| O1U123 | O1 | U12 | 3 |
| O1U133 | O1 | U13 | 3 |
| O1U233 | O1 | U23 | 3 |
| O1U114 | O1 | U11 | 4 |
| O1U224 | O1 | U22 | 4 |
| O1U334 | O1 | U33 | 4 |
| O1U124 | O1 | U12 | 4 |
| O1U134 | O1 | U13 | 4 |
| O1U234 | O1 | U23 | 4 |
| Na1U111 | Na1 | U11 | 1 |
| Na1U221 | Na1 | U22 | 1 |
| Na1U331 | Na1 | U33 | 1 |
| Na1U121 | Na1 | U12 | 1 |
| Na1U131 | Na1 | U13 | 1 |
| Na1U231 | Na1 | U23 | 1 |
| Na1U112 | Na1 | U11 | 2 |
| Na1U222 | Na1 | U22 | 2 |
| Na1U332 | Na1 | U33 | 2 |
| Na1U122 | Na1 | U12 | 2 |
| Na1U132 | Na1 | U13 | 2 |
| Na1U232 | Na1 | U23 | 2 |
| Na1U113 | Na1 | U11 | 3 |
| Na1U223 | Na1 | U22 | 3 |
| Na1U333 | Na1 | U33 | 3 |
| Na1U123 | Na1 | U12 | 3 |
| Na1U133 | Na1 | U13 | 3 |
| Na1U233 | Na1 | U23 | 3 |
| Na1U114 | Na1 | U11 | 4 |
| Na1U224 | Na1 | U22 | 4 |
| Na1U334 | Na1 | U33 | 4 |
| Na1U124 | Na1 | U12 | 4 |
| Na1U134 | Na1 | U13 | 4 |
| Na1U234 | Na1 | U23 | 4 |
ADP Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cu1U111 | 0 | 0 |
| Cu1U221 | 0 | 0 |
| Cu1U331 | 0 | 0 |
| Cu1U121 | -0.002292 | 0 |
| Cu1U131 | 0 | 0 |
| Cu1U231 | -0.001951 | 0 |
| Cu1U112 | -0.000938 | 0 |
| Cu1U222 | -0.001233 | 0 |
| Cu1U332 | -0.000409 | 0 |
| Cu1U122 | 0 | 0 |
| Cu1U132 | 0.000089 | 0 |
| Cu1U232 | 0 | 0 |
| Cu1U113 | 0 | 0 |
| Cu1U223 | 0 | 0 |
| Cu1U333 | 0 | 0 |
| Cu1U123 | 0.001153 | 0 |
| Cu1U133 | 0 | 0 |
| Cu1U233 | 0.001033 | 0 |
| Cu1U114 | -0.00367 | 0 |
| Cu1U224 | -0.00013 | 0 |
| Cu1U334 | -0.001985 | 0 |
| Cu1U124 | 0 | 0 |
| Cu1U134 | -0.001894 | 0 |
| Cu1U234 | 0 | 0 |
| O1U111 | 0 | -0.007726 |
| O1U221 | 0 | -0.001137 |
| O1U331 | 0 | 0.000357 |
| O1U121 | 0.004968 | 0 |
| O1U131 | 0 | -0.000565 |
| O1U231 | 0.002039 | 0 |
| O1U112 | 0.000177 | 0 |
| O1U222 | 0.005123 | 0 |
| O1U332 | -0.00091 | 0 |
| O1U122 | 0 | -0.000235 |
| O1U132 | 0.000452 | 0 |
| O1U232 | 0 | 0.000425 |
| O1U113 | 0 | -0.000164 |
| O1U223 | 0 | -0.000637 |
| O1U333 | 0 | -0.000686 |
| O1U123 | 0.003843 | 0 |
| O1U133 | 0 | -0.000386 |
| O1U233 | 0.000288 | 0 |
| O1U114 | -0.001578 | 0 |
| O1U224 | 0.002843 | 0 |
| O1U334 | 0.001428 | 0 |
| O1U124 | 0 | 0.000814 |
| O1U134 | 0.000707 | 0 |
| O1U234 | 0 | -0.00006 |
| Na1U111 | 0.005322 | 0 |
| Na1U221 | 0.035756 | 0 |
| Na1U331 | 0.002385 | 0 |
| Na1U121 | 0 | 0.019492 |
| Na1U131 | 0.001318 | 0 |
| Na1U231 | 0 | 0.008364 |
| Na1U112 | -0.004723 | 0 |
| Na1U222 | 0.001838 | 0 |
| Na1U332 | 0.00038 | 0 |
| Na1U122 | 0 | 0.018063 |
| Na1U132 | -0.000804 | 0 |
| Na1U232 | 0 | 0.009998 |
| Na1U113 | -0.011063 | 0 |
| Na1U223 | -0.022437 | 0 |
| Na1U333 | 0.004784 | 0 |
| Na1U123 | 0 | 0.008346 |
| Na1U133 | -0.002286 | 0 |
| Na1U233 | 0 | 0.006811 |
| Na1U114 | 0.000236 | 0 |
| Na1U224 | -0.015349 | 0 |
| Na1U334 | -0.004909 | 0 |
| Na1U124 | 0 | -619.6094 |
| Na1U134 | -0.001531 | 0 |
| Na1U234 | 0 | -6.237483 |
III
Chemical data
Structural Formula Sum: Cu1 Na1.61689 O2 [ Help ]
Formula weight: 661.6 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Superspace group name: C2/m(0β0)s0 [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,x2,-x3,1/2+x4 |
| 3 | -x1,-x2,-x3,-x4 |
| 4 | x1,-x2,x3,1/2-x4 |
| 5 | 1/2+x1,1/2+x2,x3,x4 |
| 6 | 1/2-x1,1/2+x2,-x3,1/2+x4 |
| 7 | 1/2-x1,1/2-x2,-x3,-x4 |
| 8 | 1/2+x1,1/2-x2,x3,1/2-x4 |
a: 8.23682(15) Å [ Help ]
b: 2.79269(4) Å [ Help ]
c: 5.71224(10) Å [ Help ]
α: 90 ° [ Help ]
β: 111.6696(9) ° [ Help ]
γ: 90 ° [ Help ]
Volume: 122.112(5) Å3 [ Help ]
Z: 2 [ Help ]
Modulation dimension: 1 [ Help ]
Number of subsystems: 2 [ Help ]
Subsystem code: 1 [ Help ]
Subsystem description: 1-st subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
Subsystem code: 2 [ Help ]
Subsystem description: 2-nd subsystem [ Help ]
W matrix:
| 1.0 | 0.0 | 0.0 | 0.0 |
| 0.0 | 0.0 | 0.0 | 1.0 |
| 0.0 | 0.0 | 1.0 | 0.0 |
| 0.0 | 1.0 | 0.0 | 0.0 |
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.80844(3) | 0.00000 |
μ: 32.162 mm-1 [ Help ]
Absorption correction remarks: (Jana2000; Petricek, Dusek & Palatinus, 2000) [ Help ]
Refinement details
R(all): 0.0556 [ Help ]
R(obs): 0.0546 [ Help ]
wR(obs): 0.0589 [ Help ]
wR(all): 0.0589 [ Help ]
Nb. of parameters: 37 [ Help ]
Weighting scheme: sigma [ Help ]
Δ/σ(max): 0.0489 [ Help ]
Δ/σ(mean): 0.0098 [ Help ]
Extinction method: none [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom symbol | Atom site label | Subsystem | x | y | z | Uiso/equiv | ADP type | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Occupancy | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu | Cu1 | 1 | 0 | 0.5 | 0 | 0.0024(4) | Uiso | d | . | 1 | . | . |
| O | O1 | 1 | 0.0526(3) | 0 | 0.2465(5) | 0.0208(10) | Uiso | d | . | 1 | . | . |
| Na | Na1 | 2 | -0.1566(4) | 0.25 | 0.3717(5) | 0.0183(9) | Uiso | d | . | 1 | . | . |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.00000 | 0.80844 | 0.00000 |
| 2 | 0.00000 | 1.61688 | 0.00000 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Cu1x1 | Cu1 | x | 1 |
| Cu1y1 | Cu1 | y | 1 |
| Cu1z1 | Cu1 | z | 1 |
| Cu1x2 | Cu1 | x | 2 |
| Cu1y2 | Cu1 | y | 2 |
| Cu1z2 | Cu1 | z | 2 |
| O1x1 | O1 | x | 1 |
| O1y1 | O1 | y | 1 |
| O1z1 | O1 | z | 1 |
| O1x2 | O1 | x | 2 |
| O1y2 | O1 | y | 2 |
| O1z2 | O1 | z | 2 |
| Na1x1 | Na1 | x | 1 |
| Na1y1 | Na1 | y | 1 |
| Na1z1 | Na1 | z | 1 |
| Na1x2 | Na1 | x | 2 |
| Na1y2 | Na1 | y | 2 |
| Na1z2 | Na1 | z | 2 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cu1x1 | 0 | -0.0182(3) |
| Cu1y1 | 0 | 0 |
| Cu1z1 | 0 | -0.0309(4) |
| Cu1x2 | 0 | 0 |
| Cu1y2 | 0 | -0.0023(13) |
| Cu1z2 | 0 | 0 |
| O1x1 | 0 | 0.0037(9) |
| O1y1 | 0.022(2) | 0 |
| O1z1 | 0 | 0.0180(11) |
| O1x2 | -0.0037(14) | 0 |
| O1y2 | 0 | 0.010(4) |
| O1z2 | 0.007(2) | 0 |
| Na1x1 | -0.0396(5) | 0 |
| Na1y1 | 0 | 0.1009(12) |
| Na1z1 | -0.0332(9) | 0 |
| Na1x2 | -0.0210(9) | 0 |
| Na1y2 | 0 | 0.003(2) |
| Na1z2 | -0.0190(15) | 0 |