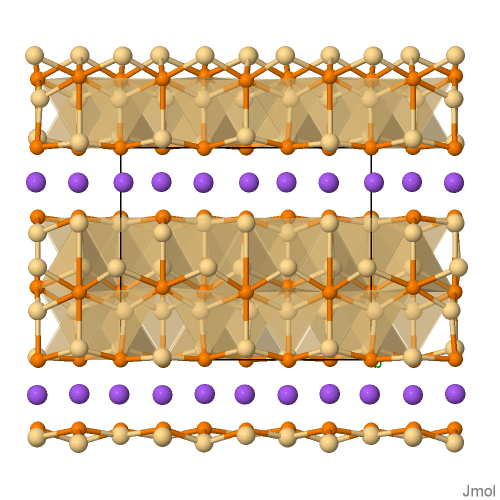
Polymorphism in Zintl-Phases ACd4Pn3: The modulated structures of NaCd4Pn3 with Pn = P, As
Authors:Grotz, Carolin; Baumgartner, Maximilian; Freitag, Katharina M.; Baumer, Franziska; Nilges, Tom
Journal:Inorganic Chemistry 55 7764-7776 (2016)
DOI:http://dx.doi.org/10.1021/acs.inorgchem.6b01233
B-IncStrDB ID: 9iBxSt5KeU3 Entry date: 2022-01-10 Last revision: 2022-02-12
Cd4Na1P3_298
Chemical data
Structural Formula Sum: Cd4 Na1 P3 [ Help ]
Formula weight: 565.6 Da [ Help ]
Crystallographic data and experimental details
Crystal system: trigonal [ Help ]
Space group name (H-M): R -3 m [ Help ]
Space group name (Hall): -R 3 2" [ Help ]
Space group nb.: 166 [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x,y,z |
| 2 | -y,x-y,z |
| 3 | -x+y,-x,z |
| 4 | y,x,-z |
| 5 | x-y,-y,-z |
| 6 | -x,-x+y,-z |
| 7 | -x,-y,-z |
| 8 | y,-x+y,-z |
| 9 | x-y,x,-z |
| 10 | -y,-x,z |
| 11 | -x+y,y,z |
| 12 | x,x-y,z |
| 13 | x+2/3,y+1/3,z+1/3 |
| 14 | -y+2/3,x-y+1/3,z+1/3 |
| 15 | -x+y+2/3,-x+1/3,z+1/3 |
| 16 | y+2/3,x+1/3,-z+1/3 |
| 17 | x-y+2/3,-y+1/3,-z+1/3 |
| 18 | -x+2/3,-x+y+1/3,-z+1/3 |
| 19 | -x+2/3,-y+1/3,-z+1/3 |
| 20 | y+2/3,-x+y+1/3,-z+1/3 |
| 21 | x-y+2/3,x+1/3,-z+1/3 |
| 22 | -y+2/3,-x+1/3,z+1/3 |
| 23 | -x+y+2/3,y+1/3,z+1/3 |
| 24 | x+2/3,x-y+1/3,z+1/3 |
| 25 | x+1/3,y+2/3,z+2/3 |
| 26 | -y+1/3,x-y+2/3,z+2/3 |
| 27 | -x+y+1/3,-x+2/3,z+2/3 |
| 28 | y+1/3,x+2/3,-z+2/3 |
| 29 | x-y+1/3,-y+2/3,-z+2/3 |
| 30 | -x+1/3,-x+y+2/3,-z+2/3 |
| 31 | -x+1/3,-y+2/3,-z+2/3 |
| 32 | y+1/3,-x+y+2/3,-z+2/3 |
| 33 | x-y+1/3,x+2/3,-z+2/3 |
| 34 | -y+1/3,-x+2/3,z+2/3 |
| 35 | -x+y+1/3,y+2/3,z+2/3 |
| 36 | x+1/3,x-y+2/3,z+2/3 |
a: 4.3445(5) Å [ Help ]
b: 4.3445(5) Å [ Help ]
c: 33.197(5) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 120 ° [ Help ]
Volume: 542.64(12) Å3 [ Help ]
Z: 3 [ Help ]
Cell determination reflection Nb.: 2479 [ Help ]
θ(min) for cell determination: 2.58 ° [ Help ]
θ(max) for cell determination: 32.17 ° [ Help ]
Cell measurement temperature: 298 K [ Help ]
μ: 12.309 mm-1 [ Help ]
Absorption correction type: gaussian [ Help ]
Absorption correction remarks: Jana2006 [ Help ]
Minimum transmission factor: 0.3914 [ Help ]
Maximum transmission factor: 0.8087 [ Help ]
Refinement details
Total nb. of reflections: 160 [ Help ]
Nb. of observed reflections: 128 [ Help ]
Intense reflections threshold: I>3σ(I) [ Help ]
Refinement based on: Fsqd [ Help ]
R(obs): 0.0369 [ Help ]
wR(obs): 0.0699 [ Help ]
R(all): 0.0492 [ Help ]
wR(all): 0.0719 [ Help ]
S(all): 1.91 [ Help ]
S(obs): 2.10 [ Help ]
Nb. of reflections: 160 [ Help ]
Nb. of parameters: 15 [ Help ]
Number of restraints: 0 [ Help ]
Number of constraints: 0 [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/(σ2(I)+0.0004I2) [ Help ]
Δ/σ(max): 0.0047 [ Help ]
Δ/σ(mean): 0.0018 [ Help ]
Δρ(max): 1.31 e_Å-3 [ Help ]
Δρ(min): -1.87 e_Å-3 [ Help ]
Extinction method: B-C type 1 Gaussian isotropic (Becker & Coppens, 1974) [ Help ]
Extinction coefficient: 490(50) [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | ADP type | Uiso/equiv | Symmetry multiplicity | Occupancy | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cd1 | Cd | 0 | 0 | 0.09768(6) | Uani | 0.0324(6) | 6 | 1 | d | . | . | . |
| P1 | P | 0 | 0 | 0.22205(12) | Uani | 0.0101(11) | 6 | 1 | d | . | . | . |
| Cd2 | Cd | 0 | 0 | 0.29950(4) | Uani | 0.0313(6) | 6 | 1 | d | . | . | . |
| Na1 | Na | 0 | 0 | 0.5 | Uani | 0.029(3) | 3 | 1 | d | . | . | . |
| P2 | P | 0 | 0 | 0 | Uani | 0.0153(16) | 3 | 1 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | Atom site symbol | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|---|
| Cd1 | Cd | 0.0120(6) | 0.0120(6) | 0.0733(13) | 0.0060(3) | 0 | 0 |
| P1 | P | 0.0095(13) | 0.0095(13) | 0.011(2) | 0.0047(7) | 0 | 0 |
| Cd2 | Cd | 0.0405(7) | 0.0405(7) | 0.0130(7) | 0.0202(4) | 0 | 0 |
| Na1 | Na | 0.027(4) | 0.027(4) | 0.034(7) | 0.013(2) | 0 | 0 |
| P2 | P | 0.0112(17) | 0.0112(17) | 0.023(3) | 0.0056(9) | 0 | 0 |
Cd4Na1P3_130_mod
Chemical data
Structural Formula Sum: Cd4 Na1 P3 [ Help ]
Formula weight: 565.6 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Superspace group name: Cm(α0γ)s [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | x1,-x2,x3,x4+1/2 |
| 3 | x1+1/2,x2+1/2,x3,x4 |
| 4 | x1+1/2,-x2+1/2,x3,x4+1/2 |
a: 7.5098(8) Å [ Help ]
b: 13.0065(10) Å [ Help ]
c: 11.2885(11) Å [ Help ]
α: 90 ° [ Help ]
β: 102.827(8) ° [ Help ]
γ: 90 ° [ Help ]
Volume: 1075.10(18) Å3 [ Help ]
Modulation dimension: 1 [ Help ]
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | -0.040000 | 0.000000 | 0.342262 |
Z: 6 [ Help ]
Cell measurement temperature: 130 K [ Help ]
μ: 12.425 mm-1 [ Help ]
Absorption correction type: gaussian [ Help ]
Absorption correction remarks: Jana2006 [ Help ]
Minimum transmission factor: 0.1598 [ Help ]
Maximum transmission factor: 0.2853 [ Help ]
Refinement details
Total nb. of reflections: 8466 [ Help ]
Nb. of observed reflections: 2895 [ Help ]
Intense reflections threshold: I>3σ(I) [ Help ]
Refinement based on: Fsqd [ Help ]
R(obs): 0.0353 [ Help ]
wR(obs): 0.0661 [ Help ]
R(all): 0.1331 [ Help ]
wR(all): 0.0927 [ Help ]
S(all): 0.89 [ Help ]
S(obs): 1.11 [ Help ]
Nb. of reflections: 8466 [ Help ]
Nb. of parameters: 211 [ Help ]
Number of restraints: 0 [ Help ]
Number of constraints: 2 [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/(σ2(I)+0.0004I2) [ Help ]
Δ/σ(max): 0.0043 [ Help ]
Δ/σ(mean): 0.0011 [ Help ]
Δρ(max): 2.18 e_Å-3 [ Help ]
Δρ(min): -1.53 e_Å-3 [ Help ]
Extinction method: B-C type 1 Gaussian isotropic (Becker & Coppens, 1974) [ Help ]
Extinction coefficient: 316(11) [ Help ]
Absolute structure remarks: 4172 of Friedel pairs used in the refinement [ Help ]
Flack parameter: 0.55(9) [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | ADP type | Uiso/equiv | Symmetry multiplicity | Occupancy | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cd1 | Cd | 0.1767(3) | 0.1661(2) | 0.0315(2) | Uani | 0.0216(6) | 4 | 1 | d | . | . | . |
| Cd2 | Cd | 0.3729(3) | 0.1670(2) | 0.6203(2) | Uani | 0.0220(6) | 4 | 1 | d | . | . | . |
| Cd3 | Cd | 0.6842(5) | 0 | 0.0525(3) | Uani | 0.0153(7) | 2 | 1 | d | . | . | . |
| Cd4 | Cd | 0.8809(6) | 0 | 0.6409(4) | Uani | 0.0229(9) | 2 | 1 | d | . | . | . |
| Cd5 | Cd | 0.0689(3) | 0.3257(2) | 0.2300(2) | Uani | 0.0214(5) | 4 | 1 | d | . | . | . |
| Cd6 | Cd | 0.4707(3) | 0.3403(3) | 0.4339(2) | Uani | 0.0195(5) | 4 | 1 | d | . | . | . |
| Cd7 | Cd | 0.0921(5) | 0 | 0.2298(4) | Uani | 0.0221(7) | 2 | 1 | d | . | . | . |
| Cd8 | Cd | 0.4926(5) | 0 | 0.4325(4) | Uani | 0.0207(7) | 2 | 1 | d | . | . | . |
| P1 | P | 0.2767(9) | 0.1665(7) | 0.3312(7) | Uani | 0.0121(11) | 4 | 1 | d | . | . | . |
| P2 | P | 0.7744(9) | 0 | 0.3197(7) | Uani | 0.009(2) | 2 | 1 | d | . | . | . |
| P3 | P | 0.0019(8) | 0.3330(5) | 0.0005(5) | Uiso | 0.0015(9) | 4 | 1 | d | . | . | . |
| P4 | P | 0.0600(10) | 0.1683(5) | 0.6720(7) | Uani | 0.0153(12) | 4 | 1 | d | . | . | . |
| P5 | P | 0.0007(15) | 0 | 0.0001(10) | Uiso | 0.0034(15) | 2 | 1 | d | . | . | . |
| P6 | P | 0.5561(16) | 0 | 0.6687(12) | Uani | 0.0163(18) | 2 | 1 | d | . | . | . |
| Na1 | Na | 0.2694(17) | 0.334(2) | 0.8353(9) | Uani | 0.0150(12) | 4 | 1 | d | . | . | . |
| Na2 | Na | 0.288(2) | 0 | 0.8353(11) | Uiso | 0.02 | 2 | 1 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | Atom site symbol | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|---|
| Cd1 | Cd | 0.0107(8) | 0.0095(8) | 0.0473(12) | 0.0004(7) | 0.0120(7) | 0.0008(8) |
| Cd2 | Cd | 0.0124(8) | 0.0126(8) | 0.0422(12) | 0.0002(7) | 0.0085(7) | 0.0010(9) |
| Cd3 | Cd | 0.0055(10) | 0.0024(10) | 0.0387(14) | 0 | 0.0068(9) | 0 |
| Cd4 | Cd | 0.0105(11) | 0.0056(12) | 0.055(2) | 0 | 0.0123(12) | 0 |
| Cd5 | Cd | 0.0250(8) | 0.0266(9) | 0.0126(6) | 0.0018(7) | 0.0041(5) | 0.0020(6) |
| Cd6 | Cd | 0.0239(8) | 0.0203(9) | 0.0136(7) | 0.0030(7) | 0.0028(6) | -0.0001(6) |
| Cd7 | Cd | 0.0256(13) | 0.0246(14) | 0.0142(10) | 0 | 0.0001(9) | 0 |
| Cd8 | Cd | 0.0193(11) | 0.0278(15) | 0.0146(10) | 0 | 0.0030(9) | 0 |
| P1 | P | 0.009(2) | 0.0096(18) | 0.0181(18) | 0.0003(7) | 0.0036(13) | -0.0008(11) |
| P2 | P | 0.009(4) | 0.007(3) | 0.011(3) | 0 | 0.004(2) | 0 |
| P4 | P | 0.0129(16) | 0.0133(15) | 0.020(2) | -0.0009(10) | 0.0042(13) | -0.0015(14) |
| P6 | P | 0.010(2) | 0.011(2) | 0.028(4) | 0 | 0.0049(19) | 0 |
| Na1 | Na | 0.0142(18) | 0.016(2) | 0.0164(18) | -0.001(3) | 0.0058(14) | 0.0002(19) |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_1 |
|---|---|
| 1 | 1 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Cd1x1 | Cd1 | x | 1 |
| Cd1y1 | Cd1 | y | 1 |
| Cd1z1 | Cd1 | z | 1 |
| Cd2x1 | Cd2 | x | 1 |
| Cd2y1 | Cd2 | y | 1 |
| Cd2z1 | Cd2 | z | 1 |
| Cd5x1 | Cd5 | x | 1 |
| Cd5y1 | Cd5 | y | 1 |
| Cd5z1 | Cd5 | z | 1 |
| Cd6x1 | Cd6 | x | 1 |
| Cd6y1 | Cd6 | y | 1 |
| Cd6z1 | Cd6 | z | 1 |
| Cd7x1 | Cd7 | x | 1 |
| Cd7y1 | Cd7 | y | 1 |
| Cd7z1 | Cd7 | z | 1 |
| Cd8x1 | Cd8 | x | 1 |
| Cd8y1 | Cd8 | y | 1 |
| Cd8z1 | Cd8 | z | 1 |
| P1x1 | P1 | x | 1 |
| P1y1 | P1 | y | 1 |
| P1z1 | P1 | z | 1 |
| P3x1 | P3 | x | 1 |
| P3y1 | P3 | y | 1 |
| P3z1 | P3 | z | 1 |
| Na1x1 | Na1 | x | 1 |
| Na1y1 | Na1 | y | 1 |
| Na1z1 | Na1 | z | 1 |
| Na2x1 | Na2 | x | 1 |
| Na2y1 | Na2 | y | 1 |
| Na2z1 | Na2 | z | 1 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cd1x1 | -0.0077(4) | 0.000(2) |
| Cd1y1 | -0.0005(4) | 0.0009(3) |
| Cd1z1 | -0.0207(4) | 0.001(6) |
| Cd2x1 | -0.0035(17) | -0.0055(11) |
| Cd2y1 | -0.0002(3) | 0.0007(3) |
| Cd2z1 | -0.009(6) | -0.021(2) |
| Cd5x1 | 0.021(2) | 0.007(6) |
| Cd5y1 | -0.0065(8) | -0.0026(19) |
| Cd5z1 | 0.0009(3) | -0.0005(4) |
| Cd6x1 | 0.015(4) | 0.016(4) |
| Cd6y1 | 0.0057(16) | 0.0056(16) |
| Cd6z1 | -0.0003(4) | 0.0010(3) |
| Cd7x1 | 0 | 0 |
| Cd7y1 | 0.0136(16) | 0.006(4) |
| Cd7z1 | 0 | 0 |
| Cd8x1 | 0 | 0 |
| Cd8y1 | -0.012(3) | -0.011(3) |
| Cd8z1 | 0 | 0 |
| P1x1 | 0.0037(9) | 0.0023(14) |
| P1y1 | -0.0007(8) | 0.0006(6) |
| P1z1 | 0.0100(18) | 0.006(3) |
| P3x1 | -0.0019(4) | -0.0007(6) |
| P3y1 | 0.0024(4) | 0.0003(7) |
| P3z1 | 0.0013(8) | -0.0027(4) |
| Na1x1 | 0.0089(19) | 0.001(4) |
| Na1y1 | -0.001(3) | -0.0038(19) |
| Na1z1 | 0.0022(13) | -0.0011(19) |
| Na2x1 | 0 | 0 |
| Na2y1 | 0.0(9) | 0.012(2) |
| Na2z1 | 0 | 0 |
Definition of the ADP Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Tensor element | Wave vector code |
|---|---|---|---|
| Cd1U111 | Cd1 | U11 | 1 |
| Cd1U221 | Cd1 | U22 | 1 |
| Cd1U331 | Cd1 | U33 | 1 |
| Cd1U121 | Cd1 | U12 | 1 |
| Cd1U131 | Cd1 | U13 | 1 |
| Cd1U231 | Cd1 | U23 | 1 |
| Cd2U111 | Cd2 | U11 | 1 |
| Cd2U221 | Cd2 | U22 | 1 |
| Cd2U331 | Cd2 | U33 | 1 |
| Cd2U121 | Cd2 | U12 | 1 |
| Cd2U131 | Cd2 | U13 | 1 |
| Cd2U231 | Cd2 | U23 | 1 |
| Cd5U111 | Cd5 | U11 | 1 |
| Cd5U221 | Cd5 | U22 | 1 |
| Cd5U331 | Cd5 | U33 | 1 |
| Cd5U121 | Cd5 | U12 | 1 |
| Cd5U131 | Cd5 | U13 | 1 |
| Cd5U231 | Cd5 | U23 | 1 |
| Cd7U111 | Cd7 | U11 | 1 |
| Cd7U221 | Cd7 | U22 | 1 |
| Cd7U331 | Cd7 | U33 | 1 |
| Cd7U121 | Cd7 | U12 | 1 |
| Cd7U131 | Cd7 | U13 | 1 |
| Cd7U231 | Cd7 | U23 | 1 |
| Na1U111 | Na1 | U11 | 1 |
| Na1U221 | Na1 | U22 | 1 |
| Na1U331 | Na1 | U33 | 1 |
| Na1U121 | Na1 | U12 | 1 |
| Na1U131 | Na1 | U13 | 1 |
| Na1U231 | Na1 | U23 | 1 |
ADP Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Cd1U111 | 0.0012(10) | 0.0017(11) |
| Cd1U221 | 0.0036(8) | 0.0000(12) |
| Cd1U331 | -0.0123(15) | 0.001(4) |
| Cd1U121 | -0.0003(10) | -0.0009(11) |
| Cd1U131 | -0.0050(13) | 0.0031(18) |
| Cd1U231 | -0.0005(15) | -0.0025(15) |
| Cd2U111 | 0.0000(12) | 0.0020(9) |
| Cd2U221 | 0.0019(16) | 0.0048(10) |
| Cd2U331 | -0.014(9) | -0.031(4) |
| Cd2U121 | -0.0002(11) | -0.0003(9) |
| Cd2U131 | -0.0040(19) | -0.0057(14) |
| Cd2U231 | -0.0030(15) | 0.0001(15) |
| Cd5U111 | 0.0103(12) | 0.001(3) |
| Cd5U221 | -0.007(2) | -0.006(2) |
| Cd5U331 | 0.0006(6) | -0.0005(6) |
| Cd5U121 | 0.002(2) | 0.0077(12) |
| Cd5U131 | -0.0013(9) | -0.0016(6) |
| Cd5U231 | 0.0005(17) | 0.0045(10) |
| Cd7U111 | 0 | 0 |
| Cd7U221 | 0 | 0 |
| Cd7U331 | 0 | 0 |
| Cd7U121 | -0.0156(15) | 0.001(4) |
| Cd7U131 | 0 | 0 |
| Cd7U231 | -0.0038(13) | 0.0003(17) |
| Na1U111 | 0.000(6) | -0.004(5) |
| Na1U221 | 0.003(6) | 0.003(5) |
| Na1U331 | -0.004(5) | -0.001(5) |
| Na1U121 | -0.002(6) | -0.001(6) |
| Na1U131 | -0.001(5) | -0.008(3) |
| Na1U231 | -0.001(6) | 0.014(6) |