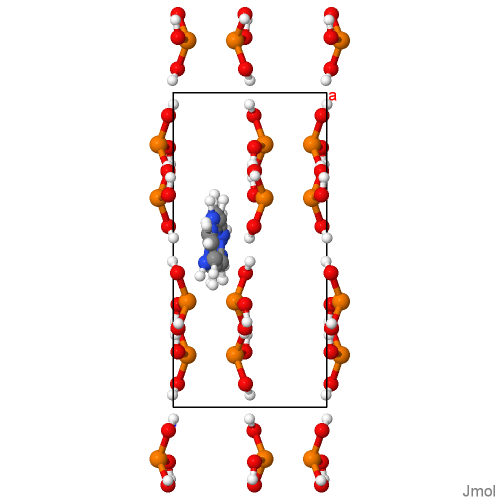
Crystal structures and phase transitions of imidazolium hypodiphosphates
Authors:Budzikur, Daria; Szklarz, Przemyslaw; Kinzhybalo, Vasyl; Slepokura, Katarzyna A.
Journal:Acta Crystallographica Section B 76 939-947 (2020)
DOI:https://doi.org/10.1107/S2052520620011439
B-IncStrDB ID: 16472EnKoLb Entry date: 2020-09-12 Last revision: 2021-12-12
I-HTP
Chemical data
Formula moiety: (C3 H5 N2), (H3 O6 P2) [ Help ]
Structural Formula Sum: C3 H8 N2 O6 P2 [ Help ]
Formula weight: 230.05 Da [ Help ]
Crystallographic data and experimental details
Crystal system: orthorhombic [ Help ]
Space group nb.: 52 [ Help ]
Space group name (H-M): P n n a [ Help ]
Space group name (Hall): -P 2a 2bc [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x, y, z |
| 2 | -x+1/2, -y, z |
| 3 | -x+1/2, y+1/2, -z+1/2 |
| 4 | x, -y+1/2, -z+1/2 |
| 5 | -x, -y, -z |
| 6 | x-1/2, y, -z |
| 7 | x-1/2, -y-1/2, z-1/2 |
| 8 | -x, y-1/2, z-1/2 |
a: 7.014(3) Å [ Help ]
b: 8.361(3) Å [ Help ]
c: 14.389(5) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 843.8(6) Å3 [ Help ]
Z: 4 [ Help ]
Cell measurement temperature: 326(2) K [ Help ]
Cell determination reflection Nb.: 4207 [ Help ]
θ(min) for cell determination: 4.70 ° [ Help ]
θ(max) for cell determination: 29.04 ° [ Help ]
μ: 0.518 mm-1 [ Help ]
Absorption correction type: analytical [ Help ]
Minimum transmission factor: 0.879 [ Help ]
Maximum transmission factor: 0.960 [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.39.46 (Rigaku Oxford Diffraction, 2018) Analytical numeric absorption correction using a multifaceted crystal model based on expressions derived by R.C. Clark & J.S. Reid. (Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887-897) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Refinement details
Total nb. of reflections: 1134 [ Help ]
Nb. of observed reflections: 863 [ Help ]
Intense reflections threshold: I > 2σ(I) [ Help ]
Proportion of Friedel related intensities: 0.000 [ Help ]
Reflection handling remarks: Reflections were merged by SHELXL according to the crystal class for the calculation of statistics and refinement. _reflns_Friedel_fraction is defined as the number of unique Friedel pairs measured divided by the number that would be possible theoretically, ignoring centric projections and systematic absences. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: w=1/[σ2(Fo2)+(0.0507P)2+0.2662P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: constr [ Help ]
Extinction method: SHELXL-2014/7 (Sheldrick 2014 [ Help ]
Extinction coefficient: 0.065(5) [ Help ]
Extinction expression: Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 [ Help ]
Nb. of reflections: 1134 [ Help ]
Nb. of parameters: 101 [ Help ]
Number of restraints: 64 [ Help ]
R(all): 0.0480 [ Help ]
R(obs): 0.0350 [ Help ]
wR(all): 0.1039 [ Help ]
wR(obs): 0.0950 [ Help ]
S(all): 1.095 [ Help ]
Restrained S(all): 1.086 [ Help ]
Δ/σ(max): 0.000 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 0.221 e_Å-3 [ Help ]
Δρ(min): -0.315 e_Å-3 [ Help ]
Δρ(rms): 0.057 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | _atom_site_site_symmetry_order | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | ADPs constraints or restraints | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P | 0.59456(8) | -0.00006(5) | 0.33491(3) | 0.0431(2) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O1 | O | 0.5287(3) | 0.07335(14) | 0.42605(9) | 0.0594(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H1 | H | 0.4973 | 0.0021 | 0.4620 | 0.089 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| O2 | O | 0.5307(2) | -0.17364(15) | 0.32531(8) | 0.0555(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2 | H | 0.5169 | -0.1954 | 0.2702 | 0.083 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| O3 | O | 0.5315(2) | 0.09995(15) | 0.25206(9) | 0.0539(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3 | H | 0.5339 | 0.1950 | 0.2661 | 0.081 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| N1A | N | 0.318(3) | 0.3850(17) | 0.4513(12) | 0.019(4) | Uiso | 0.1 | 1 | d | D | U | P | A | -1 |
| H1NA | H | 0.3469 | 0.2880 | 0.4373 | 0.023 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C2A | C | 0.276(3) | 0.497(2) | 0.3892(10) | 0.023(4) | Uiso | 0.1 | 1 | d | D | U | P | A | -1 |
| H2A | H | 0.2699 | 0.4846 | 0.3250 | 0.028 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| N3A | N | 0.245(4) | 0.630(2) | 0.4369(12) | 0.035(6) | Uiso | 0.1 | 1 | d | D | U | P | A | -1 |
| H3NA | H | 0.2140 | 0.7215 | 0.4135 | 0.042 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C4A | C | 0.270(3) | 0.5991(19) | 0.5292(11) | 0.037(5) | Uiso | 0.1 | 1 | d | D | U | P | A | -1 |
| H4A | H | 0.2621 | 0.6739 | 0.5769 | 0.045 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C5A | C | 0.309(4) | 0.442(2) | 0.5402(11) | 0.033(6) | Uiso | 0.1 | 1 | d | D | U | P | A | -1 |
| H5A | H | 0.3258 | 0.3862 | 0.5954 | 0.039 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| N1B | N | 0.3068(14) | 0.3895(9) | 0.5180(6) | 0.038(2) | Uiso | 0.2 | 1 | d | D | U | P | B | -2 |
| H1NB | H | 0.3314 | 0.3065 | 0.5512 | 0.045 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C2B | C | 0.2987(17) | 0.3952(10) | 0.4260(6) | 0.039(3) | Uiso | 0.2 | 1 | d | D | U | P | B | -2 |
| H2B | H | 0.3185 | 0.3097 | 0.3859 | 0.047 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| N3B | N | 0.2573(15) | 0.5438(10) | 0.4012(6) | 0.034(2) | Uiso | 0.2 | 1 | d | D | U | P | B | -2 |
| H3NB | H | 0.2395 | 0.5753 | 0.3450 | 0.041 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C4B | C | 0.247(2) | 0.6397(12) | 0.4777(6) | 0.058(4) | Uiso | 0.2 | 1 | d | D | U | P | B | -2 |
| H4B | H | 0.2295 | 0.7499 | 0.4794 | 0.069 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C5B | C | 0.2687(15) | 0.5398(10) | 0.5504(6) | 0.033(3) | Uiso | 0.2 | 1 | d | D | U | P | B | -2 |
| H5B | H | 0.2591 | 0.5685 | 0.6127 | 0.039 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| N1C | N | 0.212(2) | 0.5834(16) | 0.5399(7) | 0.021(2) | Uiso | 0.2 | 1 | d | D | U | P | C | -3 |
| H1NC | H | 0.1745 | 0.6461 | 0.5836 | 0.025 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C2C | C | 0.254(2) | 0.4299(17) | 0.5503(8) | 0.045(5) | Uiso | 0.2 | 1 | d | D | U | P | C | -3 |
| H2C | H | 0.2476 | 0.3719 | 0.6054 | 0.054 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| N3C | N | 0.3059(18) | 0.3738(11) | 0.4679(7) | 0.027(2) | Uiso | 0.2 | 1 | d | D | U | P | C | -3 |
| H3NC | H | 0.3395 | 0.2769 | 0.4564 | 0.032 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C4C | C | 0.2976(15) | 0.4949(13) | 0.4046(7) | 0.025(2) | Uiso | 0.2 | 1 | d | D | U | P | C | -3 |
| H4C | H | 0.3281 | 0.4874 | 0.3418 | 0.031 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C5C | C | 0.237(3) | 0.6272(14) | 0.4492(9) | 0.034(4) | Uiso | 0.2 | 1 | d | D | U | P | C | -3 |
| H5C | H | 0.2172 | 0.7279 | 0.4235 | 0.041 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| P1 | 0.0726(4) | 0.0295(3) | 0.0271(3) | -0.00153(16) | 0.00558(18) | -0.00982(19) |
| O1 | 0.1093(12) | 0.0349(7) | 0.0339(7) | -0.0041(5) | 0.0159(7) | 0.0028(7) |
| O2 | 0.0904(10) | 0.0317(7) | 0.0444(7) | -0.0047(5) | 0.0149(7) | -0.0191(7) |
| O3 | 0.0817(9) | 0.0471(7) | 0.0328(6) | 0.0053(5) | -0.0079(6) | -0.0095(7) |
I_RTP
Chemical data
Formula moiety: (C3 H5 N2), (H3 O6 P2) [ Help ]
Structural Formula Sum: C3 H8 N2 O6 P2 [ Help ]
Formula weight: 230.05 Da [ Help ]
Crystallographic data and experimental details
Crystal system: orthorhombic [ Help ]
Space group nb.: 52 [ Help ]
Space group name (H-M): P n n a [ Help ]
Space group name (Hall): -P 2a 2bc [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x, y, z |
| 2 | -x+1/2, -y, z |
| 3 | -x+1/2, y+1/2, -z+1/2 |
| 4 | x, -y+1/2, -z+1/2 |
| 5 | -x, -y, -z |
| 6 | x-1/2, y, -z |
| 7 | x-1/2, -y-1/2, z-1/2 |
| 8 | -x, y-1/2, z-1/2 |
a: 6.962(3) Å [ Help ]
b: 8.368(3) Å [ Help ]
c: 14.388(4) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 838.2(5) Å3 [ Help ]
Z: 4 [ Help ]
Cell measurement temperature: 291(2) K [ Help ]
Cell determination reflection Nb.: 4526 [ Help ]
θ(min) for cell determination: 4.7210 ° [ Help ]
θ(max) for cell determination: 29.0850 ° [ Help ]
μ: 0.522 mm-1 [ Help ]
Absorption correction type: analytical [ Help ]
Minimum transmission factor: 0.868 [ Help ]
Maximum transmission factor: 0.957 [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.39.46 (Rigaku Oxford Diffraction, 2018) Analytical numeric absorption correction using a multifaceted crystal model based on expressions derived by R.C. Clark & J.S. Reid. (Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887-897) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Refinement details
Total nb. of reflections: 1128 [ Help ]
Nb. of observed reflections: 901 [ Help ]
Intense reflections threshold: I > 2σ(I) [ Help ]
Proportion of Friedel related intensities: 0.000 [ Help ]
Reflection handling remarks: Reflections were merged by SHELXL according to the crystal class for the calculation of statistics and refinement. _reflns_Friedel_fraction is defined as the number of unique Friedel pairs measured divided by the number that would be possible theoretically, ignoring centric projections and systematic absences. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: w=1/[σ2(Fo2)+(0.0419P)2+0.4756P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: constr [ Help ]
Extinction method: SHELXL-2014/7 (Sheldrick 2015) [ Help ]
Extinction coefficient: 0.065(4) [ Help ]
Extinction expression: Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 [ Help ]
Nb. of reflections: 1128 [ Help ]
Nb. of parameters: 101 [ Help ]
Number of restraints: 35 [ Help ]
R(all): 0.0456 [ Help ]
R(obs): 0.0351 [ Help ]
wR(all): 0.0949 [ Help ]
wR(obs): 0.0865 [ Help ]
S(all): 1.050 [ Help ]
Restrained S(all): 1.035 [ Help ]
Δ/σ(max): 0.000 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 0.235 e_Å-3 [ Help ]
Δρ(min): -0.395 e_Å-3 [ Help ]
Δρ(rms): 0.057 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | _atom_site_site_symmetry_order | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | ADPs constraints or restraints | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P | 0.59322(8) | 0.00009(5) | 0.33478(3) | 0.0410(2) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O1 | O | 0.5268(3) | 0.07328(15) | 0.42589(9) | 0.0537(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H1 | H | 0.514694 | 0.002730 | 0.465006 | 0.081 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| O2 | O | 0.5286(3) | -0.17341(16) | 0.32510(9) | 0.0524(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2 | H | 0.522141 | -0.196980 | 0.269866 | 0.079 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| O3 | O | 0.5298(2) | 0.10050(16) | 0.25196(9) | 0.0509(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3 | H | 0.536109 | 0.195536 | 0.265515 | 0.076 | Uiso | 0.5 | 1 | calc | R | U | P | ? | ? |
| N1A | N | 0.315(4) | 0.3899(19) | 0.4357(10) | 0.018(5) | Uiso | 0.1 | 1 | d | D | ? | P | A | -1 |
| H1A | H | 0.341530 | 0.301228 | 0.408235 | 0.022 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C2A | C | 0.262(4) | 0.519(2) | 0.3881(10) | 0.014(4) | Uiso | 0.1 | 1 | d | D | ? | P | A | -1 |
| H2A | H | 0.240136 | 0.530347 | 0.324739 | 0.017 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| N3A | N | 0.249(5) | 0.626(2) | 0.4558(12) | 0.042(11) | Uiso | 0.1 | 1 | d | D | ? | P | A | -1 |
| H3A | H | 0.222872 | 0.725672 | 0.446907 | 0.051 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C4A | C | 0.282(4) | 0.5602(19) | 0.5414(12) | 0.027(6) | Uiso | 0.1 | 1 | d | D | ? | P | A | -1 |
| H4A | H | 0.273958 | 0.613810 | 0.597862 | 0.033 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| C5A | C | 0.327(3) | 0.405(2) | 0.5304(10) | 0.010(3) | Uiso | 0.1 | 1 | d | D | ? | P | A | -1 |
| H5A | H | 0.357571 | 0.328163 | 0.574925 | 0.012 | Uiso | 0.1 | 1 | calc | R | U | P | A | -1 |
| N1B | N | 0.285(2) | 0.3941(11) | 0.5211(7) | 0.036(3) | Uiso | 0.2 | 1 | d | D | ? | P | B | -2 |
| H1B | H | 0.309717 | 0.313912 | 0.556397 | 0.044 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C2B | C | 0.268(2) | 0.3814(9) | 0.4293(7) | 0.029(3) | Uiso | 0.2 | 1 | d | D | ? | P | B | -2 |
| H2B | H | 0.286994 | 0.292095 | 0.391904 | 0.035 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| N3B | N | 0.2177(18) | 0.5286(9) | 0.4046(6) | 0.024(3) | Uiso | 0.2 | 1 | d | D | ? | P | B | -2 |
| H3B | H | 0.201627 | 0.558654 | 0.348010 | 0.028 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C4B | C | 0.194(2) | 0.6268(14) | 0.4798(7) | 0.036(3) | Uiso | 0.2 | 1 | d | D | ? | P | B | -2 |
| H4B | H | 0.143740 | 0.729692 | 0.479366 | 0.043 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| C5B | C | 0.259(3) | 0.5459(12) | 0.5546(8) | 0.038(4) | Uiso | 0.2 | 1 | d | D | ? | P | B | -2 |
| H5B | H | 0.280296 | 0.583857 | 0.614431 | 0.046 | Uiso | 0.2 | 1 | calc | R | U | P | B | -2 |
| N1C | N | 0.212(3) | 0.5622(16) | 0.5424(8) | 0.027(3) | Uiso | 0.2 | 1 | d | D | ? | P | C | -3 |
| H1C | H | 0.165905 | 0.609116 | 0.590781 | 0.033 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C2C | C | 0.274(2) | 0.4121(16) | 0.5447(8) | 0.018(3) | Uiso | 0.2 | 1 | d | D | ? | P | C | -3 |
| H2C | H | 0.288262 | 0.344285 | 0.595466 | 0.022 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| N3C | N | 0.311(3) | 0.3842(15) | 0.4554(8) | 0.022(3) | Uiso | 0.2 | 1 | d | D | ? | P | C | -3 |
| H3C | H | 0.343391 | 0.292508 | 0.433545 | 0.026 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C4C | C | 0.290(2) | 0.5190(15) | 0.4027(8) | 0.027(3) | Uiso | 0.2 | 1 | d | D | ? | P | C | -3 |
| H4C | H | 0.315575 | 0.527623 | 0.339451 | 0.032 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
| C5C | C | 0.225(3) | 0.6374(14) | 0.4580(10) | 0.021(4) | Uiso | 0.2 | 1 | d | D | ? | P | C | -3 |
| H5C | H | 0.196349 | 0.742762 | 0.442819 | 0.025 | Uiso | 0.2 | 1 | calc | R | U | P | C | -3 |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| P1 | 0.0685(4) | 0.0304(3) | 0.0240(3) | -0.00231(17) | 0.00466(19) | -0.0146(2) |
| O1 | 0.1012(12) | 0.0316(7) | 0.0283(7) | -0.0036(5) | 0.0124(7) | -0.0010(7) |
| O2 | 0.0860(11) | 0.0316(7) | 0.0394(7) | -0.0052(5) | 0.0148(7) | -0.0218(7) |
| O3 | 0.0763(9) | 0.0488(8) | 0.0276(6) | 0.0044(5) | -0.0064(6) | -0.0128(7) |
I_LTP
Chemical data
Formula moiety: (C3 H5 N2), (H3 O6 P2) [ Help ]
Structural Formula Sum: C3 H8 N2 O6 P2 [ Help ]
Formula weight: 230.05 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Space group nb.: 14 [ Help ]
Space group name (H-M): P 21/n 1 1 [ Help ]
Space group name (Hall): -P 2xn [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x, y, z |
| 2 | x+1/2, -y+1/2, -z+1/2 |
| 3 | -x, -y, -z |
| 4 | -x-1/2, y-1/2, z-1/2 |
a: 13.673(2) Å [ Help ]
b: 8.396(2) Å [ Help ]
c: 14.317(3) Å [ Help ]
α: 90.57(2) ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 1643.5(6) Å3 [ Help ]
Z: 8 [ Help ]
Cell measurement temperature: 100(2) K [ Help ]
Cell determination reflection Nb.: 6843 [ Help ]
θ(min) for cell determination: 2.84 ° [ Help ]
θ(max) for cell determination: 29.26 ° [ Help ]
μ: 0.532 mm-1 [ Help ]
Absorption correction type: analytical [ Help ]
Minimum transmission factor: 0.868 [ Help ]
Maximum transmission factor: 0.952 [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.39.46 (Rigaku Oxford Diffraction, 2018) Analytical numeric absorption correction using a multifaceted crystal model based on expressions derived by R.C. Clark & J.S. Reid. (Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887-897) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Refinement details
Total nb. of reflections: 4008 [ Help ]
Nb. of observed reflections: 3358 [ Help ]
Intense reflections threshold: I > 2σ(I) [ Help ]
Proportion of Friedel related intensities: 0.000 [ Help ]
Reflection handling remarks: Reflections were merged by SHELXL according to the crystal class for the calculation of statistics and refinement. _reflns_Friedel_fraction is defined as the number of unique Friedel pairs measured divided by the number that would be possible theoretically, ignoring centric projections and systematic absences. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: w=1/[σ2(Fo2)+(0.0355P)2+1.1991P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: mixed [ Help ]
Extinction method: SHELXL-2014/7 (Sheldrick 2015) [ Help ]
Extinction coefficient: 0.0049(3) [ Help ]
Extinction expression: Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 [ Help ]
Nb. of reflections: 4008 [ Help ]
Nb. of parameters: 245 [ Help ]
Number of restraints: 5 [ Help ]
R(all): 0.0411 [ Help ]
R(obs): 0.0318 [ Help ]
wR(all): 0.0833 [ Help ]
wR(obs): 0.0774 [ Help ]
S(all): 1.042 [ Help ]
Restrained S(all): 1.041 [ Help ]
Δ/σ(max): 0.000 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 0.430 e_Å-3 [ Help ]
Δρ(min): -0.472 e_Å-3 [ Help ]
Δρ(rms): 0.081 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | _atom_site_site_symmetry_order | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | ADPs constraints or restraints | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1A | P | 0.28745(3) | 0.01736(5) | 0.33674(3) | 0.00790(11) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| P2A | P | 0.44733(3) | 0.01986(5) | 0.33894(3) | 0.00804(11) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O1A | O | 0.25123(9) | 0.09237(13) | 0.42644(8) | 0.0109(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O2A | O | 0.25705(9) | 0.10470(14) | 0.24807(8) | 0.0108(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O3A | O | 0.25329(9) | -0.15954(13) | 0.33107(9) | 0.0120(3) | Uani | 1 | 1 | d | D | ? | ? | ? | ? |
| H3A | H | 0.2625 | -0.2064 | 0.2797 | 0.066(10) | Uiso | 1 | 1 | d | DR | ? | ? | ? | ? |
| O4A | O | 0.47699(9) | 0.19232(13) | 0.33225(8) | 0.0107(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O5A | O | 0.48196(9) | -0.06658(14) | 0.42585(8) | 0.0122(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H5A | H | 0.5000 | 0.0000 | 0.5000 | 0.082(16) | Uiso | 1 | 2 | d | S | ? | P | ? | ? |
| O6A | O | 0.48091(9) | -0.07278(14) | 0.24998(8) | 0.0117(3) | Uani | 1 | 1 | d | D | ? | ? | ? | ? |
| H6A | H | 0.4847 | -0.1724 | 0.2537 | 0.069(10) | Uiso | 1 | 1 | d | DR | ? | ? | ? | ? |
| P1B | P | 0.31216(3) | 0.50861(5) | 0.16782(3) | 0.00846(11) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| P2B | P | 0.47219(3) | 0.51986(5) | 0.16949(3) | 0.00814(11) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O1B | O | 0.27275(9) | 0.67343(13) | 0.18213(8) | 0.0114(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O2B | O | 0.28276(9) | 0.39351(14) | 0.24509(8) | 0.0121(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2B | H | 0.275(3) | 0.263(4) | 0.242(2) | 0.079(11) | Uiso | 1 | 1 | d | ? | ? | ? | ? | ? |
| O3B | O | 0.28407(9) | 0.43198(14) | 0.07219(8) | 0.0124(3) | Uani | 1 | 1 | d | D | ? | ? | ? | ? |
| H3B | H | 0.2763 | 0.4955 | 0.0279 | 0.057(9) | Uiso | 1 | 1 | d | DR | ? | ? | ? | ? |
| O4B | O | 0.50262(9) | 0.62797(14) | 0.24819(8) | 0.0114(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O5B | O | 0.50603(9) | 0.57711(14) | 0.07341(8) | 0.0113(3) | Uani | 1 | 1 | d | D | ? | ? | ? | ? |
| H5B | H | 0.5088 | 0.5162 | 0.0266 | 0.017 | Uiso | 0.5 | 1 | d | DR | U | P | ? | ? |
| O6B | O | 0.50977(9) | 0.34540(14) | 0.18031(9) | 0.0117(3) | Uani | 1 | 1 | d | D | ? | ? | ? | ? |
| H6B | H | 0.5014 | 0.3034 | 0.2327 | 0.074(11) | Uiso | 1 | 1 | d | DR | ? | ? | ? | ? |
| N1A | N | 0.15762(11) | 0.38999(17) | 0.45119(10) | 0.0123(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H1NA | H | 0.1801 | 0.2971 | 0.4318 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| N3A | N | 0.10820(11) | 0.63204(17) | 0.45115(11) | 0.0125(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3NA | H | 0.0923 | 0.7277 | 0.4318 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C2A | C | 0.14201(13) | 0.5153(2) | 0.39745(13) | 0.0136(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2AA | H | 0.1531 | 0.5203 | 0.3320 | 0.016 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C4A | C | 0.10181(13) | 0.5810(2) | 0.54167(12) | 0.0121(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H4A | H | 0.0797 | 0.6414 | 0.5940 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C5A | C | 0.13314(13) | 0.4282(2) | 0.54205(13) | 0.0127(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H5AA | H | 0.1374 | 0.3604 | 0.5946 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| N1B | N | 0.40281(11) | 0.40332(17) | 0.46499(10) | 0.0119(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H1NB | H | 0.4246 | 0.3331 | 0.4239 | 0.014 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| N3B | N | 0.35492(11) | 0.50904(17) | 0.59381(11) | 0.0137(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3NB | H | 0.3394 | 0.5212 | 0.6531 | 0.016 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C2B | C | 0.38577(13) | 0.3749(2) | 0.55451(13) | 0.0141(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2BA | H | 0.3942 | 0.2758 | 0.5850 | 0.017 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C4B | C | 0.35081(13) | 0.6264(2) | 0.52792(13) | 0.0127(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H4B | H | 0.3306 | 0.7335 | 0.5376 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C5B | C | 0.38112(13) | 0.5599(2) | 0.44651(13) | 0.0128(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H5BA | H | 0.3864 | 0.6114 | 0.3879 | 0.015 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| P1A | 0.0103(2) | 0.0068(2) | 0.0067(2) | 0.00033(14) | 0.00073(17) | -0.00010(16) |
| P2A | 0.0102(2) | 0.0067(2) | 0.0072(2) | 0.00017(15) | 0.00022(17) | 0.00057(15) |
| O1A | 0.0137(6) | 0.0100(6) | 0.0091(6) | -0.0007(4) | 0.0026(5) | 0.0018(5) |
| O2A | 0.0140(7) | 0.0102(6) | 0.0082(6) | 0.0019(4) | -0.0019(5) | -0.0020(5) |
| O3A | 0.0164(7) | 0.0081(6) | 0.0117(6) | 0.0000(4) | 0.0024(5) | -0.0023(5) |
| O4A | 0.0134(6) | 0.0081(6) | 0.0107(6) | -0.0007(4) | 0.0003(5) | -0.0011(5) |
| O5A | 0.0172(7) | 0.0104(6) | 0.0091(6) | 0.0002(4) | -0.0032(5) | 0.0022(5) |
| O6A | 0.0167(7) | 0.0088(6) | 0.0096(6) | -0.0014(4) | 0.0022(5) | 0.0014(5) |
| P1B | 0.0107(2) | 0.0068(2) | 0.0079(2) | -0.00017(15) | 0.00009(17) | -0.00058(16) |
| P2B | 0.0108(2) | 0.0068(2) | 0.0069(2) | -0.00094(15) | 0.00065(17) | -0.00064(16) |
| O1B | 0.0145(6) | 0.0083(6) | 0.0114(6) | -0.0002(4) | -0.0005(5) | 0.0014(5) |
| O2B | 0.0162(7) | 0.0099(6) | 0.0103(6) | 0.0010(4) | 0.0025(5) | -0.0013(5) |
| O3B | 0.0186(7) | 0.0095(6) | 0.0089(6) | -0.0005(4) | -0.0030(5) | -0.0018(5) |
| O4B | 0.0147(6) | 0.0095(6) | 0.0101(6) | -0.0022(4) | -0.0015(5) | -0.0011(5) |
| O5B | 0.0165(7) | 0.0096(6) | 0.0078(6) | -0.0001(4) | 0.0023(5) | -0.0029(5) |
| O6B | 0.0151(7) | 0.0084(6) | 0.0117(6) | 0.0005(5) | 0.0022(5) | 0.0010(5) |
| N1A | 0.0116(7) | 0.0094(7) | 0.0160(8) | -0.0032(5) | -0.0008(6) | 0.0021(6) |
| N3A | 0.0122(8) | 0.0092(7) | 0.0164(8) | 0.0030(5) | -0.0004(6) | 0.0013(6) |
| C2A | 0.0112(9) | 0.0177(9) | 0.0119(9) | 0.0000(7) | 0.0005(7) | 0.0002(7) |
| C4A | 0.0126(9) | 0.0137(8) | 0.0100(9) | -0.0021(6) | 0.0014(7) | -0.0009(7) |
| C5A | 0.0126(9) | 0.0136(8) | 0.0119(9) | 0.0029(6) | -0.0020(7) | -0.0007(7) |
| N1B | 0.0120(8) | 0.0105(7) | 0.0130(8) | -0.0042(5) | 0.0013(6) | 0.0005(6) |
| N3B | 0.0160(8) | 0.0161(8) | 0.0089(7) | -0.0014(6) | 0.0030(6) | 0.0000(6) |
| C2B | 0.0132(9) | 0.0122(8) | 0.0170(9) | 0.0020(7) | -0.0012(7) | 0.0003(7) |
| C4B | 0.0121(9) | 0.0101(8) | 0.0159(9) | -0.0008(6) | -0.0014(7) | 0.0012(7) |
| C5B | 0.0148(9) | 0.0128(8) | 0.0109(9) | 0.0024(6) | -0.0009(7) | -0.0012(7) |
II
Chemical data
Formula moiety: 2(C3 H5 N2), (H2 O6 P2) [ Help ]
Structural Formula Sum: C6 H12 N4 O6 P2 [ Help ]
Formula weight: 298.14 Da [ Help ]
Absolute configuration: ad [ Help ]
Crystallographic data and experimental details
Crystal system: tetragonal [ Help ]
Space group nb.: 96 [ Help ]
Space group name (H-M): P 43 21 2 [ Help ]
Space group name (Hall): P 4nw 2abw [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x, y, z |
| 2 | -x, -y, z+1/2 |
| 3 | -y+1/2, x+1/2, z+3/4 |
| 4 | y+1/2, -x+1/2, z+1/4 |
| 5 | -x+1/2, y+1/2, -z+3/4 |
| 6 | x+1/2, -y+1/2, -z+1/4 |
| 7 | y, x, -z |
| 8 | -y, -x, -z+1/2 |
a: 8.966(2) Å [ Help ]
b: 8.966(2) Å [ Help ]
c: 15.462(4) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 1243.0(6) Å3 [ Help ]
Z: 4 [ Help ]
Cell measurement temperature: 110(2) K [ Help ]
Cell determination reflection Nb.: 5341 [ Help ]
θ(min) for cell determination: 2.61 ° [ Help ]
θ(max) for cell determination: 28.44 ° [ Help ]
μ: 0.376 mm-1 [ Help ]
Absorption correction type: analytical [ Help ]
Minimum transmission factor: 0.867 [ Help ]
Maximum transmission factor: 0.983 [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.39.46 (Rigaku Oxford Diffraction, 2018) Analytical numeric absorption correction using a multifaceted crystal model based on expressions derived by R.C. Clark & J.S. Reid. (Clark, R. C. & Reid, J. S. (1995). Acta Cryst. A51, 887-897) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Refinement details
Total nb. of reflections: 1550 [ Help ]
Nb. of observed reflections: 1444 [ Help ]
Intense reflections threshold: I > 2σ(I) [ Help ]
Proportion of Friedel related intensities: 0.601 [ Help ]
Fraction of Friedel pairs out to θ(max)): 0.937 [ Help ]
Fraction of Friedel pairs out to θ(full): 1.000 [ Help ]
Reflection handling remarks: Reflections were merged by SHELXL according to the crystal class for the calculation of statistics and refinement. _reflns_Friedel_fraction is defined as the number of unique Friedel pairs measured divided by the number that would be possible theoretically, ignoring centric projections and systematic absences. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: w=1/[σ2(Fo2)+(0.0313P)2+0.3895P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: mixed [ Help ]
Extinction method: none [ Help ]
Absolute structure remarks: Flack x determined using 519 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons, Flack and Wagner, Acta Cryst. B69 (2013) 249-259). [ Help ]
Flack parameter: 0.03(4) [ Help ]
Nb. of reflections: 1550 [ Help ]
Nb. of parameters: 94 [ Help ]
R(all): 0.0326 [ Help ]
R(obs): 0.0284 [ Help ]
wR(all): 0.0646 [ Help ]
wR(obs): 0.0631 [ Help ]
S(all): 1.084 [ Help ]
Restrained S(all): 1.084 [ Help ]
Δ/σ(max): 0.001 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 0.297 e_Å-3 [ Help ]
Δρ(min): -0.298 e_Å-3 [ Help ]
Δρ(rms): 0.048 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | _atom_site_site_symmetry_order | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | ADPs constraints or restraints | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P | 0.31970(6) | 0.24790(6) | 0.56384(3) | 0.01203(14) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O1 | O | 0.29901(18) | 0.08245(17) | 0.57054(11) | 0.0214(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O2 | O | 0.47546(16) | 0.30850(17) | 0.57853(10) | 0.0161(3) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| O3 | O | 0.21351(18) | 0.33388(19) | 0.62805(10) | 0.0189(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3 | H | 0.131(4) | 0.288(4) | 0.635(2) | 0.054(11) | Uiso | 1 | 1 | d | ? | ? | ? | ? | ? |
| N1 | N | 0.4969(2) | 0.6045(2) | 0.61837(13) | 0.0187(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H1N | H | 0.493(4) | 0.518(4) | 0.606(2) | 0.038(9) | Uiso | 1 | 1 | d | ? | ? | ? | ? | ? |
| N3 | N | 0.4378(2) | 0.8365(2) | 0.61309(13) | 0.0189(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H3N | H | 0.401(4) | 0.911(4) | 0.599(2) | 0.046(10) | Uiso | 1 | 1 | d | ? | ? | ? | ? | ? |
| C2 | C | 0.4252(3) | 0.7072(2) | 0.57305(15) | 0.0195(5) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H2 | H | 0.3733 | 0.6906 | 0.5204 | 0.023 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C4 | C | 0.5189(2) | 0.8159(3) | 0.68759(16) | 0.0196(5) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H4 | H | 0.5442 | 0.8902 | 0.7288 | 0.024 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
| C5 | C | 0.5557(2) | 0.6700(2) | 0.69101(15) | 0.0187(4) | Uani | 1 | 1 | d | ? | ? | ? | ? | ? |
| H5 | H | 0.6115 | 0.6220 | 0.7352 | 0.022 | Uiso | 1 | 1 | calc | R | U | ? | ? | ? |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| P1 | 0.0132(3) | 0.0100(3) | 0.0128(2) | 0.0010(2) | -0.0022(2) | -0.0007(2) |
| O1 | 0.0240(9) | 0.0120(7) | 0.0281(9) | 0.0037(7) | -0.0086(7) | -0.0015(6) |
| O2 | 0.0146(8) | 0.0151(7) | 0.0188(8) | 0.0010(6) | -0.0040(6) | -0.0008(6) |
| O3 | 0.0188(8) | 0.0208(9) | 0.0170(8) | -0.0033(6) | 0.0040(6) | -0.0051(7) |
| N1 | 0.0191(10) | 0.0135(10) | 0.0236(10) | -0.0021(8) | 0.0005(8) | 0.0007(7) |
| N3 | 0.0217(10) | 0.0127(10) | 0.0225(10) | 0.0001(8) | -0.0044(8) | 0.0030(8) |
| C2 | 0.0203(11) | 0.0181(11) | 0.0200(11) | -0.0025(8) | -0.0032(9) | 0.0009(8) |
| C4 | 0.0201(11) | 0.0172(11) | 0.0216(11) | -0.0027(10) | -0.0037(10) | 0.0001(9) |
| C5 | 0.0181(10) | 0.0177(11) | 0.0201(10) | 0.0003(10) | -0.0023(9) | -0.0006(9) |