NaCeP2Se6, Cu0.4Ce1.2P2Se6, Ce4(P2Se6)3, and the Incommensurately Modulated AgCeP2Se6: New Selenophosphates Featuring the Ethane-Like [P2Se6]4- Anion
Authors:Aitken, Jennifer A.; Evain, Michel; Iordanidis, Lykourgos; Kanatzidis, Mercouri G.
Journal:Inorganic Chemistry 41 180-191 (2002)
DOI:https://doi.org/10.1021/ic010618p
B-IncStrDB ID: 10482EmQKuS Entry date: 2015-02-19 Last revision: 2024-01-02
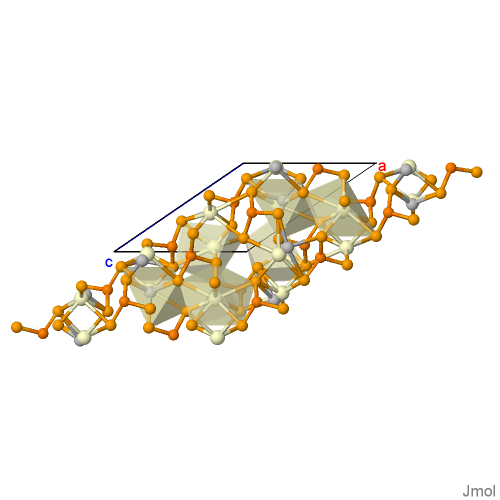
AgCePSe6
Chemical data
Structural Formula Sum: Ag1 Ce1 P2 Se6 [ Help ]
Formula weight: 783.7 Da [ Help ]
Crystallographic data and experimental details
a: 9.9557(7) Å [ Help ]
b: 7.4728(5) Å [ Help ]
c: 11.7383(8) Å [ Help ]
α: 90 ° [ Help ]
β: 145.598(5) ° [ Help ]
γ: 90 ° [ Help ]
Volume: 493.41(8) Å3 [ Help ]
Radiation used for cell measurement: Mo Kα [ Help ]
Cell determination reflection Nb.: 903 [ Help ]
Cell measurement temperature: 273 K [ Help ]
θ(max) for cell determination: 27.97 ° [ Help ]
θ(min) for cell determination: 3.17 ° [ Help ]
λ for cell measurement: 0.71073 Å [ Help ]
Modulation dimension: 1 [ Help ]
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.312 | 0 | 0.412 |
Z: 2 [ Help ]
Crystal system: monoclinic [ Help ]
Superspace group name (ITC): P21/c(α0γ) [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,1/2+x2,1/2-x3,-x4 |
| 3 | -x1,-x2,-x3,-x4 |
| 4 | x1,1/2-x2,1/2+x3,x4 |
Absorption correction type: none [ Help ]
μ: 28.917 mm-1 [ Help ]
Minimum transmission factor: 1 [ Help ]
Maximum transmission factor: 1 [ Help ]
Refinement details
h(min): -13 [ Help ]
h(max): 7 [ Help ]
k(max): 9 [ Help ]
l(max): 15 [ Help ]
m1(min): -3 [ Help ]
m1(max): 3 [ Help ]
Intense reflections threshold: 3 [ Help ]
Total nb. of reflections: 2539 [ Help ]
Nb. of observed reflections: 2539 [ Help ]
Refinement based on: Fsqd [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/(σ2(I)+0.001024I2) [ Help ]
R(obs): 0.0458 [ Help ]
wR(obs): 0.0903 [ Help ]
R(all): 0.0458 [ Help ]
wR(all): 0.0903 [ Help ]
Nb. of reflections: 2539 [ Help ]
Nb. of parameters: 207 [ Help ]
S(all): 1.56 [ Help ]
S(obs): 1.56 [ Help ]
Δ/σ(max): 0.0002 [ Help ]
Δ/σ(mean): 0.0000 [ Help ]
Δρ(max): 3.94 e_Å-3 [ Help ]
Δρ(min): -2.51 e_Å-3 [ Help ]
Extinction method: B-C type 1 Lorentzian isotropic [ Help ]
Extinction coefficient: 0.058075 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | ADP type | x | y | z | Uiso/equiv | Symmetry multiplicity | Occupancy |
|---|---|---|---|---|---|---|---|---|
| Ag | Ag | Uani | 0.282(6) | 0.5577(4) | 0.5299(12) | 0.042(4) | 4 | 1 |
| Ce | Ce | Uani | 0.28671(18) | 0.6106(2) | 0.5411(2) | 0.015(2) | 4 | 1 |
| P | P | Uani | 0.3732(3) | 0.1072(2) | 0.4367(2) | 0.0137(18) | 4 | 1 |
| Se1 | Se | Uani | 0.12048(14) | -0.01825(12) | 0.37571(13) | 0.0201(15) | 4 | 1 |
| Se2 | Se | Uani | 0.60461(13) | 0.30711(14) | 0.67695(12) | 0.0198(9) | 4 | 1 |
| Se3 | Se | Uani | 0.21244(14) | 0.21199(12) | 0.1695(11) | 0.0191(11) | 4 | 1 |
ADP components: (Show/hide table) [ Help ]
| Atom site label | Atom site symbol | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|---|
| Ag | Ag | 0.0407(14) | 0.051(2) | 0.032(2) | 0.0025(14) | 0.0293(17) | -0.0083(19) |
| Ce | Ce | 0.0179(6) | 0.018(12) | 0.0161(11) | -0.0002(6) | 0.0156(8) | 0.0021(8) |
| P | P | 0.0129(8) | 0.016(11) | 0.0122(8) | 0.0011(7) | 0.0104(7) | 0.0009(7) |
| Se1 | Se | 0.0223(6) | 0.0211(8) | 0.0238(6) | 0.0007(4) | 0.0203(6) | 0.0031(4) |
| Se2 | Se | 0.0166(3) | 0.0171(5) | 0.0164(4) | -0.0011(4) | 0.0118(3) | -0.0036(4) |
| Se3 | Se | 0.0185(5) | 0.0227(5) | 0.0141(4) | -0.0005(4) | 0.013(5) | 0.0019(4) |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.312 | 0 | 0.412 |
| 2 | 0.624 | 0 | 0.824 |
| 3 | 0.936 | 0 | 1.236 |
Definition of the occupation Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Wave vector code |
|---|---|---|
| Ago1 | Ag | 1 |
| Ceo1 | Ce | 1 |
Occupation Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Ago1 | 0 | 0.5238(18) |
| Ceo1 | 0 | 1.0299(19) |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Agx1 | Ag | x | 1 |
| Agy1 | Ag | y | 1 |
| Agz1 | Ag | z | 1 |
| Agx2 | Ag | x | 2 |
| Agy2 | Ag | y | 2 |
| Agz2 | Ag | z | 2 |
| Cex1 | Ce | x | 1 |
| Cey1 | Ce | y | 1 |
| Cez1 | Ce | z | 1 |
| Cex2 | Ce | x | 2 |
| Cey2 | Ce | y | 2 |
| Cez2 | Ce | z | 2 |
| Px1 | P | x | 1 |
| Py1 | P | y | 1 |
| Pz1 | P | z | 1 |
| Px2 | P | x | 2 |
| Py2 | P | y | 2 |
| Pz2 | P | z | 2 |
| Se1x1 | Se1 | x | 1 |
| Se1y1 | Se1 | y | 1 |
| Se1z1 | Se1 | z | 1 |
| Se1x2 | Se1 | x | 2 |
| Se1y2 | Se1 | y | 2 |
| Se1z2 | Se1 | z | 2 |
| Se1x3 | Se1 | x | 3 |
| Se1y3 | Se1 | y | 3 |
| Se1z3 | Se1 | z | 3 |
| Se2x1 | Se2 | x | 1 |
| Se2y1 | Se2 | y | 1 |
| Se2z1 | Se2 | z | 1 |
| Se2x2 | Se2 | x | 2 |
| Se2y2 | Se2 | y | 2 |
| Se2z2 | Se2 | z | 2 |
| Se2x3 | Se2 | x | 3 |
| Se2y3 | Se2 | y | 3 |
| Se2z3 | Se2 | z | 3 |
| Se3x1 | Se3 | x | 1 |
| Se3y1 | Se3 | y | 1 |
| Se3z1 | Se3 | z | 1 |
| Se3x2 | Se3 | x | 2 |
| Se3y2 | Se3 | y | 2 |
| Se3z2 | Se3 | z | 2 |
| Se3x3 | Se3 | x | 3 |
| Se3y3 | Se3 | y | 3 |
| Se3z3 | Se3 | z | 3 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Agx1 | -0.0108(11) | 0.0114(6) |
| Agy1 | 0.0154(6) | -0.0321(5) |
| Agz1 | -0.041(2) | 0.0335(11) |
| Agx2 | 0.0151(15) | 0.0028(8) |
| Agy2 | 0.0123(11) | -0.012(7) |
| Agz2 | 0.0277(15) | 0.0215(9) |
| Cex1 | 0.0008(4) | 0.0173(2) |
| Cey1 | -0.0028(3) | -0.017(3) |
| Cez1 | 0.0018(4) | 0.0195(2) |
| Cex2 | -0.0044(12) | -0.0011(8) |
| Cey2 | 0.0076(10) | 0.0024(6) |
| Cez2 | -0.0112(9) | -0.0013(7) |
| Px1 | -0.005(4) | -0.003(4) |
| Py1 | -0.0111(4) | -0.0273(4) |
| Pz1 | -0.0082(3) | 0.0054(3) |
| Px2 | 0.0072(11) | 0.0051(10) |
| Py2 | 0.0034(7) | 0.0003(7) |
| Pz2 | 0(9) | -0.0007(9) |
| Se1x1 | -0.0347(2) | 0.0053(2) |
| Se1y1 | -0.01278(18) | -0.0279(2) |
| Se1z1 | -0.0324(2) | 0.0091(2) |
| Se1x2 | 0.0194(5) | -0.0075(6) |
| Se1y2 | -0.0052(3) | -0.0053(4) |
| Se1z2 | 0.0142(4) | -0.0042(5) |
| Se1x3 | -0.0064(10) | 0.0036(11) |
| Se1y3 | 0.009(7) | 0.0085(6) |
| Se1z3 | -0.0071(9) | 0.0016(10) |
| Se2x1 | 0.0029(2) | -0.0051(2) |
| Se2y1 | 0.0293(2) | -0.0278(2) |
| Se2z1 | -0.01662(18) | 0.0017(19) |
| Se2x2 | -0.0042(5) | 0.005(5) |
| Se2y2 | -0.008(3) | 0.0053(4) |
| Se2z2 | 0.0057(4) | -0.0026(5) |
| Se2x3 | -0.0014(10) | 0.0029(11) |
| Se2y3 | 0.0125(6) | 0.004(6) |
| Se2z3 | -0.0062(8) | 0.0003(9) |
| Se3x1 | 0.0137(2) | -0.0296(2) |
| Se3y1 | -0.0255(17) | 0.00004(18) |
| Se3z1 | 0.00727(17) | -0.00399(17) |
| Se3x2 | -0.0009(6) | -0.0018(5) |
| Se3y2 | 0.0026(4) | 0.0009(3) |
| Se3z2 | -0.0004(5) | 0.0022(4) |
| Se3x3 | -0.0055(11) | -0.0124(10) |
| Se3y3 | 0.0068(6) | 0.0019(7) |
| Se3z3 | -0.005(11) | -0.0034(10) |
Definition of the ADP Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Tensor element | Wave vector code |
|---|---|---|---|
| AgU111 | Ag | U11 | 1 |
| AgU221 | Ag | U22 | 1 |
| AgU331 | Ag | U33 | 1 |
| AgU121 | Ag | U12 | 1 |
| AgU131 | Ag | U13 | 1 |
| AgU231 | Ag | U23 | 1 |
| CeU111 | Ce | U11 | 1 |
| CeU221 | Ce | U22 | 1 |
| CeU331 | Ce | U33 | 1 |
| CeU121 | Ce | U12 | 1 |
| CeU131 | Ce | U13 | 1 |
| CeU231 | Ce | U23 | 1 |
| Se1U111 | Se1 | U11 | 1 |
| Se1U221 | Se1 | U22 | 1 |
| Se1U331 | Se1 | U33 | 1 |
| Se1U121 | Se1 | U12 | 1 |
| Se1U131 | Se1 | U13 | 1 |
| Se1U231 | Se1 | U23 | 1 |
| Se2U111 | Se2 | U11 | 1 |
| Se2U221 | Se2 | U22 | 1 |
| Se2U331 | Se2 | U33 | 1 |
| Se2U121 | Se2 | U12 | 1 |
| Se2U131 | Se2 | U13 | 1 |
| Se2U231 | Se2 | U23 | 1 |
| Se3U111 | Se3 | U11 | 1 |
| Se3U221 | Se3 | U22 | 1 |
| Se3U331 | Se3 | U33 | 1 |
| Se3U121 | Se3 | U12 | 1 |
| Se3U131 | Se3 | U13 | 1 |
| Se3U231 | Se3 | U23 | 1 |
ADP Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| AgU111 | 0(2) | -0.0014(16) |
| AgU221 | -0.016(17) | -0.005(2) |
| AgU331 | 0.001(3) | -0.009(2) |
| AgU121 | -0.0106(13) | 0.0123(14) |
| AgU131 | 0.005(2) | -0.004(2) |
| AgU231 | -0.0064(18) | 0.016(2) |
| CeU111 | 0.0011(9) | 0.0019(6) |
| CeU221 | 0.0049(12) | -0.0015(12) |
| CeU331 | 0.008(10) | -0.0048(9) |
| CeU121 | -0.0059(7) | 0.0027(6) |
| CeU131 | 0.0035(9) | -0.0011(7) |
| CeU231 | -0.0099(8) | 0.0055(8) |
| Se1U111 | -0.0104(11) | 0.0024(9) |
| Se1U221 | 0.0001(8) | -0.0014(11) |
| Se1U331 | -0.0064(11) | 0.0005(8) |
| Se1U121 | 0.0015(7) | 0.0079(8) |
| Se1U131 | -0.0089(11) | 0.0014(8) |
| Se1U231 | 0.0027(7) | 0.0069(8) |
| Se2U111 | -0.0002(6) | -0.0004(6) |
| Se2U221 | -0.0016(11) | 0.0042(9) |
| Se2U331 | 0.0017(7) | 0.0009(6) |
| Se2U121 | -0.0026(7) | 0.0019(6) |
| Se2U131 | 0.0021(5) | -0.0015(5) |
| Se2U231 | 0.0001(8) | -0.0025(6) |
| Se3U111 | 0.0043(9) | 0.0015(6) |
| Se3U221 | -0.0048(8) | -0.0022(9) |
| Se3U331 | 0.0017(6) | 0.0025(6) |
| Se3U121 | 0.0001(6) | 0.0004(6) |
| Se3U131 | 0.0029(7) | 0.0024(6) |
| Se3U231 | -0.0007(5) | -0.0015(6) |
NaCeP2Se6
Chemical data
Structural Formula Sum: Ce Na P2 Se6 [ Help ]
Formula weight: 698.81 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Space group name (H-M): P 21/c [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x,y,z |
| 2 | -x,y+1/2,-z+1/2 |
| 3 | -x,-y,-z |
| 4 | x,-y-1/2,z-1/2 |
a: 12.1422(2) Å [ Help ]
b: 7.69820(10) Å [ Help ]
c: 11.7399(2) Å [ Help ]
α: 90.00 ° [ Help ]
β: 111.5450(10) ° [ Help ]
γ: 90.00 ° [ Help ]
Volume: 1020.69(3) Å3 [ Help ]
Z: 4 [ Help ]
Cell measurement temperature: 293(2) K [ Help ]
Cell determination reflection Nb.: 4391 [ Help ]
θ(min) for cell determination: 1.8 ° [ Help ]
θ(max) for cell determination: 26.99 ° [ Help ]
μ: 26.138 mm-1 [ Help ]
Absorption correction type: empirical [ Help ]
Minimum transmission factor: 0.439830 [ Help ]
Maximum transmission factor: 1.000000 [ Help ]
Refinement details
Total nb. of reflections: 2083 [ Help ]
Nb. of observed reflections: 1863 [ Help ]
Intense reflections threshold: >2s(I) [ Help ]
Refinement remarks: Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: calc w=1/[σ2(Fo2)+(0.0549P)2+0.0000P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: mixed [ Help ]
Extinction method: none [ Help ]
Nb. of reflections: 2083 [ Help ]
Nb. of parameters: 91 [ Help ]
Number of restraints: 0 [ Help ]
R(all): 0.0393 [ Help ]
R(obs): 0.0352 [ Help ]
wR(all): 0.0895 [ Help ]
wR(obs): 0.0879 [ Help ]
S(all): 1.074 [ Help ]
Restrained S(all): 1.074 [ Help ]
Δ/σ(max): 0.002 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 2.039 e_Å-3 [ Help ]
Δρ(min): -2.913 e_Å-3 [ Help ]
Δρ(rms): 0.433 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | Symmetry multiplicity | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ce1 | Ce | 0.34312(3) | 0.10588(5) | 0.58852(4) | 0.00420(14) | Uani | 1 | 1 | d | . | . | . |
| Se1 | Se | 0.08634(6) | 0.00172(9) | 0.34318(7) | 0.00733(18) | Uani | 1 | 1 | d | . | . | . |
| Se2 | Se | 0.11320(6) | 0.20147(9) | 0.05944(7) | 0.00601(18) | Uani | 1 | 1 | d | . | . | . |
| Se3 | Se | 0.79687(6) | 0.20738(9) | 0.37312(7) | 0.00650(18) | Uani | 1 | 1 | d | . | . | . |
| Se4 | Se | 0.31010(6) | 0.17940(9) | 0.85407(6) | 0.00567(18) | Uani | 1 | 1 | d | . | . | . |
| Se5 | Se | 0.44848(6) | 0.02568(9) | 0.18201(7) | 0.00556(18) | Uani | 1 | 1 | d | . | . | . |
| Se6 | Se | 0.59375(6) | 0.19512(9) | 0.56352(6) | 0.00493(18) | Uani | 1 | 1 | d | . | . | . |
| P1 | P | 0.19343(16) | 0.1123(2) | 0.24898(17) | 0.0039(4) | Uani | 1 | 1 | d | . | . | . |
| P2 | P | 0.68140(16) | 0.0991(2) | 0.75217(17) | 0.0034(4) | Uani | 1 | 1 | d | . | . | . |
| Na1 | Na | 0.8598(3) | 0.1071(4) | 0.1233(3) | 0.0124(7) | Uani | 1 | 1 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| Ce1 | 0.0023(2) | 0.0046(2) | 0.0056(2) | 0.00012(15) | 0.00128(17) | 0.00032(14) |
| Se1 | 0.0063(4) | 0.0081(4) | 0.0096(4) | 0.0012(3) | 0.0054(3) | 0.0001(3) |
| Se2 | 0.0023(4) | 0.0084(4) | 0.0054(4) | 0.0016(3) | -0.0008(3) | 0.0000(3) |
| Se3 | 0.0025(4) | 0.0063(4) | 0.0087(4) | 0.0027(3) | -0.0003(3) | -0.0002(2) |
| Se4 | 0.0039(4) | 0.0060(4) | 0.0057(4) | 0.0001(3) | 0.0002(3) | 0.0012(3) |
| Se5 | 0.0022(4) | 0.0075(4) | 0.0074(4) | 0.0017(3) | 0.0022(3) | 0.0011(3) |
| Se6 | 0.0031(4) | 0.0063(4) | 0.0042(4) | 0.0007(3) | -0.0001(3) | 0.0011(2) |
| P1 | 0.0016(9) | 0.0047(9) | 0.0039(9) | 0.0004(7) | -0.0006(7) | 0.0003(7) |
| P2 | 0.0015(9) | 0.0043(9) | 0.0041(9) | 0.0009(7) | 0.0008(7) | 0.0009(6) |
| Na1 | 0.0069(15) | 0.0117(16) | 0.0156(16) | 0.0012(13) | 0.0006(13) | -0.0012(12) |
Cu0-4Ce1-2P2Se6
Chemical data
Structural Formula Sum: Ce1.20 Cu0.40 P2 Se6 [ Help ]
Formula weight: 729.26 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Space group name (H-M): P 21/c [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x,y,z |
| 2 | -x,y+1/2,-z+1/2 |
| 3 | -x,-y,-z |
| 4 | x,-y-1/2,z-1/2 |
a: 12.0409(14) Å [ Help ]
b: 7.6418(8) Å [ Help ]
c: 11.6999(13) Å [ Help ]
α: 90.00 ° [ Help ]
β: 111.269(2) ° [ Help ]
γ: 90.00 ° [ Help ]
Volume: 1003.23(19) Å3 [ Help ]
Z: 4 [ Help ]
Cell measurement temperature: 293(2) K [ Help ]
Cell determination reflection Nb.: 1566 [ Help ]
θ(min) for cell determination: 3.23 ° [ Help ]
θ(max) for cell determination: 27.12 ° [ Help ]
μ: 28.258 mm-1 [ Help ]
Absorption correction type: empirical [ Help ]
Minimum transmission factor: 0.694397 [ Help ]
Maximum transmission factor: 1.000000 [ Help ]
Refinement details
Total nb. of reflections: 2150 [ Help ]
Nb. of observed reflections: 1586 [ Help ]
Intense reflections threshold: >2σ(I) [ Help ]
Refinement remarks: Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: calc w=1/[σ2(Fo2)+(0.0530P)2+0.0000P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: mixed [ Help ]
Extinction method: none [ Help ]
Nb. of reflections: 2150 [ Help ]
Nb. of parameters: 102 [ Help ]
Number of restraints: 1 [ Help ]
R(all): 0.0631 [ Help ]
R(obs): 0.0430 [ Help ]
wR(all): 0.1022 [ Help ]
wR(obs): 0.0968 [ Help ]
S(all): 0.975 [ Help ]
Restrained S(all): 0.975 [ Help ]
Δ/σ(max): 0.001 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 2.160 e_Å-3 [ Help ]
Δρ(min): -1.675 e_Å-3 [ Help ]
Δρ(rms): 0.399 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | Symmetry multiplicity | Coords from (d)iffraction or (c)alculated | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ce1 | Ce | 0.34521(6) | 0.10723(8) | 0.59066(5) | 0.01718(18) | Uani | 1 | 1 | d | . | . | . |
| Se1 | Se | 0.07631(11) | -0.00855(14) | 0.33234(10) | 0.0211(3) | Uani | 1 | 1 | d | . | . | . |
| Se2 | Se | 0.11195(11) | 0.20214(15) | 0.05700(9) | 0.0198(3) | Uani | 1 | 1 | d | . | . | . |
| Se3 | Se | 0.79822(11) | 0.20691(15) | 0.37616(10) | 0.0244(3) | Uani | 1 | 1 | d | . | . | . |
| Se4 | Se | 0.30382(11) | 0.17902(14) | 0.85441(9) | 0.0181(3) | Uani | 1 | 1 | d | . | . | . |
| Se5 | Se | 0.44748(10) | 0.02295(14) | 0.18031(9) | 0.0162(3) | Uani | 1 | 1 | d | . | . | . |
| Se6 | Se | 0.59711(10) | 0.19570(13) | 0.56492(9) | 0.0158(3) | Uani | 1 | 1 | d | . | . | . |
| P1 | P | 0.1898(2) | 0.1141(3) | 0.2457(2) | 0.0125(6) | Uani | 1 | 1 | d | . | . | . |
| P2 | P | 0.6834(3) | 0.0995(3) | 0.7531(2) | 0.0118(5) | Uani | 1 | 1 | d | . | . | . |
| Ce2 | Ce | 0.8563(3) | 0.1145(5) | 0.1148(3) | 0.0285(10) | Uani | 0.1984(17) | 1 | d | P | . | . |
| Cu1 | Cu | 0.9350(4) | 0.2224(5) | 0.2860(4) | 0.0382(15) | Uani | 0.405(5) | 1 | d | P | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| Ce1 | 0.0211(4) | 0.0151(3) | 0.0180(3) | 0.0004(2) | 0.0103(3) | 0.0003(3) |
| Se1 | 0.0260(7) | 0.0193(6) | 0.0253(6) | 0.0046(4) | 0.0179(5) | 0.0031(5) |
| Se2 | 0.0197(7) | 0.0227(6) | 0.0128(5) | 0.0034(4) | 0.0011(4) | 0.0002(5) |
| Se3 | 0.0188(7) | 0.0188(6) | 0.0297(6) | 0.0122(5) | 0.0016(5) | -0.0023(5) |
| Se4 | 0.0235(7) | 0.0153(5) | 0.0153(5) | 0.0021(4) | 0.0069(5) | 0.0044(5) |
| Se5 | 0.0142(6) | 0.0181(5) | 0.0186(5) | 0.0047(4) | 0.0088(5) | 0.0029(5) |
| Se6 | 0.0197(7) | 0.0148(5) | 0.0117(5) | 0.0015(4) | 0.0044(4) | 0.0028(5) |
| P1 | 0.0123(15) | 0.0139(13) | 0.0116(11) | 0.0002(10) | 0.0048(11) | 0.0022(11) |
| P2 | 0.0136(15) | 0.0107(12) | 0.0120(11) | 0.0016(10) | 0.0056(11) | 0.0027(11) |
| Ce2 | 0.027(2) | 0.029(2) | 0.0307(19) | -0.0004(15) | 0.0117(16) | 0.0004(16) |
| Cu1 | 0.044(3) | 0.028(2) | 0.055(3) | 0.0121(19) | 0.033(2) | 0.0117(19) |
Ce1-33P2Se6
Chemical data
Structural Formula Sum: Ce1.33 P2 Se6 [ Help ]
Formula weight: 722.06 Da [ Help ]
Crystallographic data and experimental details
Crystal system: monoclinic [ Help ]
Space group name (H-M): P 21/c [ Help ]
Symmetry operations of the space group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x,y,z |
| 2 | -x,y+1/2,-z+1/2 |
| 3 | -x,-y,-z |
| 4 | x,-y-1/2,z-1/2 |
a: 6.8057(5) Å [ Help ]
b: 22.9685(15) Å [ Help ]
c: 11.7226(8) Å [ Help ]
α: 90.00 ° [ Help ]
β: 124.0960(10) ° [ Help ]
γ: 90.00 ° [ Help ]
Volume: 1517.44(18) Å3 [ Help ]
Z: 6 [ Help ]
Cell measurement temperature: 293(2) K [ Help ]
Cell determination reflection Nb.: 3195 [ Help ]
θ(min) for cell determination: 1.77 ° [ Help ]
θ(max) for cell determination: 27.01 ° [ Help ]
μ: 27.774 mm-1 [ Help ]
Absorption correction type: empirical [ Help ]
Minimum transmission factor: 0.335814 [ Help ]
Maximum transmission factor: 1.000000 [ Help ]
Refinement details
Total nb. of reflections: 3285 [ Help ]
Nb. of observed reflections: 1822 [ Help ]
Intense reflections threshold: >2σ(I) [ Help ]
Refinement remarks: Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. [ Help ]
Refinement based on: Fsqd [ Help ]
Matrix type used for the least-squares: full [ Help ]
Weighting scheme: calc [ Help ]
Weighting scheme remarks: calc w=1/[σ2(Fo2)+(0.0609P)2+0.0000P] where P=(Fo2+2Fc2)/3 [ Help ]
Refinement of hydrogen atoms: mixed [ Help ]
Extinction method: none [ Help ]
Nb. of reflections: 3285 [ Help ]
Nb. of parameters: 139 [ Help ]
Number of restraints: 1 [ Help ]
R(all): 0.0955 [ Help ]
R(obs): 0.0555 [ Help ]
wR(all): 0.1474 [ Help ]
wR(obs): 0.1350 [ Help ]
S(all): 0.912 [ Help ]
Restrained S(all): 0.912 [ Help ]
Δ/σ(max): 0.000 [ Help ]
Δ/σ(mean): 0.000 [ Help ]
Δρ(max): 2.316 e_Å-3 [ Help ]
Δρ(min): -2.138 e_Å-3 [ Help ]
Δρ(rms): 0.430 e_Å-3 [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | Uiso/equiv | ADP type | Occupancy | Symmetry multiplicity | Coords from (d)iffraction or (c)alculated | Occ constraints or restraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ce1 | Ce | 0.7935(6) | 0.13020(17) | 0.7496(3) | 0.0525(12) | Uani | 0.345(3) | 1 | d | P | . | . |
| Ce2 | Ce | 0.1996(2) | 0.20631(4) | 0.24594(11) | 0.0181(3) | Uani | 0.778(2) | 1 | d | P | . | . |
| Ce3 | Ce | 0.78861(17) | 0.03602(3) | 0.25224(9) | 0.0133(3) | Uani | 0.875(3) | 1 | d | P | . | . |
| Se1 | Se | 0.2935(2) | 0.06602(6) | 0.96022(14) | 0.0152(3) | Uani | 1 | 1 | d | . | . | . |
| Se2 | Se | 0.0964(3) | 0.22883(6) | 0.93713(14) | 0.0183(4) | Uani | 1 | 1 | d | . | . | . |
| Se3 | Se | 0.8909(2) | 0.06417(6) | 0.55389(14) | 0.0156(3) | Uani | 1 | 1 | d | . | . | . |
| Se4 | Se | 0.2708(3) | 0.10024(6) | 0.44563(14) | 0.0173(3) | Uani | 1 | 1 | d | . | . | . |
| Se5 | Se | 0.7059(3) | 0.23254(6) | 0.53932(14) | 0.0179(4) | Uani | 1 | 1 | d | . | . | . |
| Se6 | Se | 0.6184(3) | 0.16312(6) | 0.23134(13) | 0.0165(3) | Uani | 1 | 1 | d | . | . | . |
| Se7 | Se | 0.3834(3) | 0.17049(6) | 0.76306(14) | 0.0185(4) | Uani | 1 | 1 | d | . | . | . |
| Se8 | Se | 0.8931(2) | 0.10216(6) | 0.06052(14) | 0.0150(3) | Uani | 1 | 1 | d | . | . | . |
| Se9 | Se | 0.6245(3) | 0.00539(6) | 0.73349(14) | 0.0179(3) | Uani | 1 | 1 | d | . | . | . |
| P1 | P | 0.1262(6) | 0.13190(16) | 0.5622(3) | 0.0117(7) | Uani | 1 | 1 | d | . | . | . |
| P2 | P | 0.1273(6) | 0.03520(14) | 0.0652(3) | 0.0101(7) | Uani | 1 | 1 | d | . | . | . |
| P3 | P | 0.8719(7) | 0.20279(14) | 0.4352(4) | 0.0120(8) | Uani | 1 | 1 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | U11 | U22 | U33 | U23 | U13 | U12 |
|---|---|---|---|---|---|---|
| Ce1 | 0.041(2) | 0.063(2) | 0.046(2) | -0.0061(18) | 0.0197(17) | 0.0036(19) |
| Ce2 | 0.0216(7) | 0.0151(6) | 0.0197(6) | -0.0004(4) | 0.0128(5) | -0.0001(4) |
| Ce3 | 0.0148(6) | 0.0099(4) | 0.0154(5) | -0.0011(4) | 0.0085(4) | -0.0017(4) |
| Se1 | 0.0163(8) | 0.0148(7) | 0.0222(7) | -0.0022(5) | 0.0154(7) | -0.0039(6) |
| Se2 | 0.0195(9) | 0.0153(7) | 0.0251(8) | 0.0020(6) | 0.0155(7) | 0.0055(6) |
| Se3 | 0.0140(8) | 0.0135(7) | 0.0212(7) | 0.0016(5) | 0.0111(7) | -0.0009(6) |
| Se4 | 0.0178(9) | 0.0162(7) | 0.0257(8) | -0.0021(6) | 0.0169(7) | 0.0005(6) |
| Se5 | 0.0167(9) | 0.0204(8) | 0.0242(8) | -0.0003(6) | 0.0162(7) | 0.0026(6) |
| Se6 | 0.0160(8) | 0.0148(7) | 0.0142(7) | -0.0016(5) | 0.0059(7) | 0.0041(6) |
| Se7 | 0.0165(9) | 0.0167(7) | 0.0156(7) | -0.0028(5) | 0.0048(7) | 0.0016(6) |
| Se8 | 0.0148(8) | 0.0115(7) | 0.0224(7) | -0.0018(5) | 0.0126(7) | 0.0004(6) |
| Se9 | 0.0154(8) | 0.0209(8) | 0.0144(7) | 0.0023(6) | 0.0066(7) | -0.0025(6) |
| P1 | 0.0108(19) | 0.0107(16) | 0.0150(16) | 0.0004(14) | 0.0082(15) | -0.0006(15) |
| P2 | 0.0079(19) | 0.0091(16) | 0.0137(17) | -0.0011(13) | 0.0064(16) | -0.0033(14) |
| P3 | 0.010(2) | 0.0108(18) | 0.0164(18) | 0.0010(13) | 0.0080(17) | 0.0006(13) |